Arabinogalactan endo-1,4-beta-galactosidase
Asperfillus aculeatus beta-1,4-Galactanase (AAGAL) is an enzyme involved in pectin degradation. It belongs to the glycoside hydrolase family 53 (GH-53) in clan GH-A. It catalyses the endohydrolysis of beta-1,4-linked galactan and type I arabinogalactan to galactose and galactose oligomers.
Galactan and arabinogalactan are components of pectin which attaches to the C4 position of rhamnose. They forms the side-chains of the 'hairy' region of pectin. Degradation and modification of the galatan and arabinogalactan side chain has many industrial applications, so AAGAL has potential industrial uses.
Reference Protein and Structure
- Sequence
-
P48842
 (3.2.1.89)
(3.2.1.89)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Aspergillus aculeatus (Fungus)

- PDB
-
1fhl
- CRYSTAL STRUCTURE OF BETA-1,4-GALACTANASE FROM ASPERGILLUS ACULEATUS AT 293K
(2.3 Å)



- Catalytic CATH Domains
-
3.20.20.80
 (see all for 1fhl)
(see all for 1fhl)
Enzyme Reaction (EC:3.2.1.89)
Enzyme Mechanism
Introduction
Glu136 protonates the glycosidic oxygen while Glu246 acts as a nucleophilic to attack the anomeric carbon to form a covalent enzyme-substrate intermediate. The deprotonated Glu136 then activates a water molecule to hydrolyse the covalent enzyme-substrate intermediate. Arg45 hydrogen bonds with Glu246 to ensure that it is deprotonated even at relatively low pH value to allow it to act as a catalytic nucleophile.
Catalytic Residues Roles
| UniProt | PDB* (1fhl) | ||
| Glu152 | Glu136A | Protonates glycosyl oxygen to facilitate nucleophilic attack on the anomeric carbon resulting in the glycosyl-enzyme intermediate. Then activates water to allow hydrolysis of the intermediate. | increase nucleophilicity, activator, proton acceptor, proton donor |
| Glu262 | Glu246A | Attacks anomeric carbon to form the glycosyl-enzyme intermediate which subsequently is hydrolysed to give the products. | covalently attached, hydrogen bond acceptor, nucleofuge, nucleophile |
| Arg61 | Arg45A | Hydrogen bonds with Glu 246 and alters the pKa of Glu 246 to keep it deprotonated even at relatively low pH value to allow it to act as a nucleophile to attack the anomeric carbon of the glycosidic bond. | modifies pKa, increase nucleophilicity, hydrogen bond donor, electrostatic stabiliser |
Chemical Components
proton transfer, intermediate formation, bimolecular nucleophilic substitution, overall product formed, overall reactant used, intermediate terminated, hydrolysis, native state of enzyme regeneratedReferences
- Ryttersgaard C et al. (2002), Biochemistry, 41, 15135-15143. Aspergillus aculeatusβ-1,4-Galactanase: Substrate Recognition and Relations to Other Glycoside Hydrolases in Clan GH-A†. DOI:10.1021/bi026238c. PMID:12484750.

Step 1. Glu246 performs a nucleophilic attack on the glycosidic linkage while Glu136 protonates the leaving group. This leads to an enzyme-substrate intermediate being formed.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu246A | covalently attached |
| Arg45A | modifies pKa, increase nucleophilicity, electrostatic stabiliser, hydrogen bond donor |
| Glu246A | hydrogen bond acceptor, nucleophile |
| Glu136A | proton donor |
Chemical Components
proton transfer, intermediate formation, ingold: bimolecular nucleophilic substitution, overall product formed, overall reactant used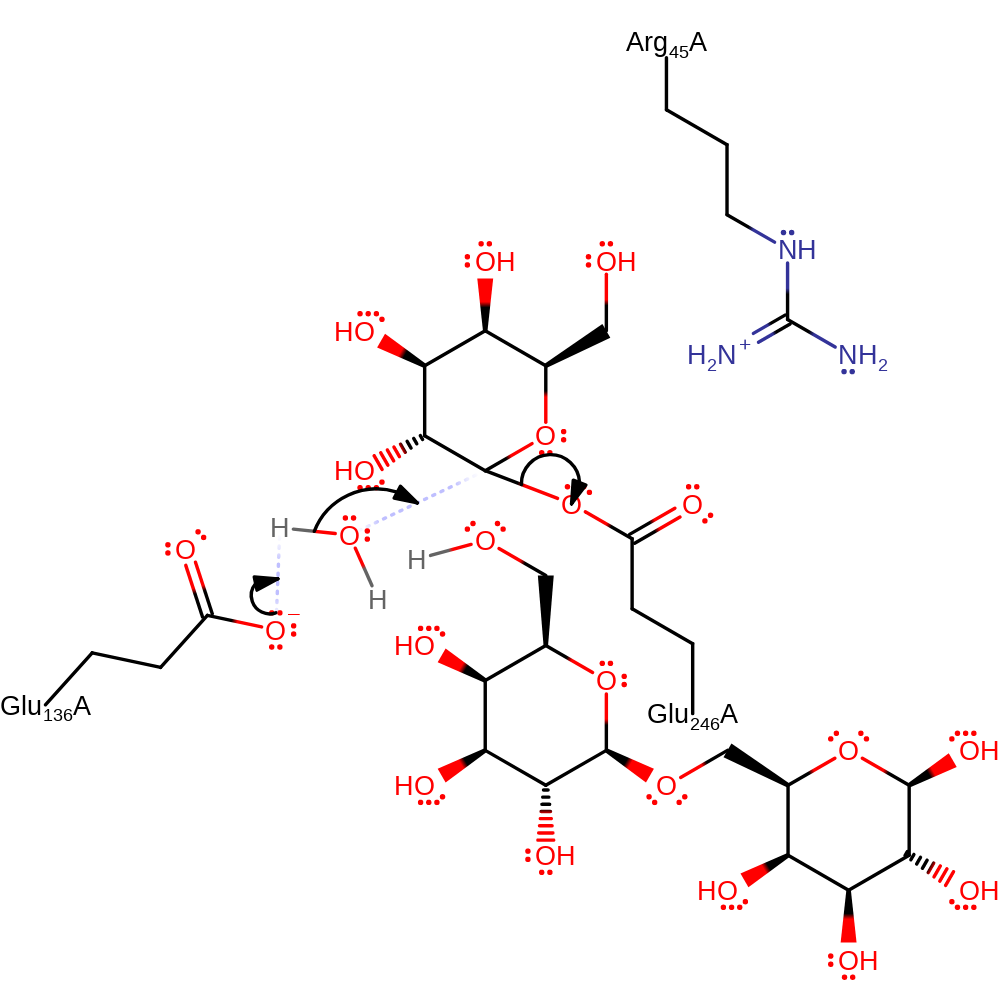
Step 2. Glu136 activates a water for nucleophilic attack, which leads to the intermediate being hydrolysed and the active site being regenerated.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu136A | increase nucleophilicity, activator |
| Glu246A | nucleofuge |
| Glu136A | proton acceptor |




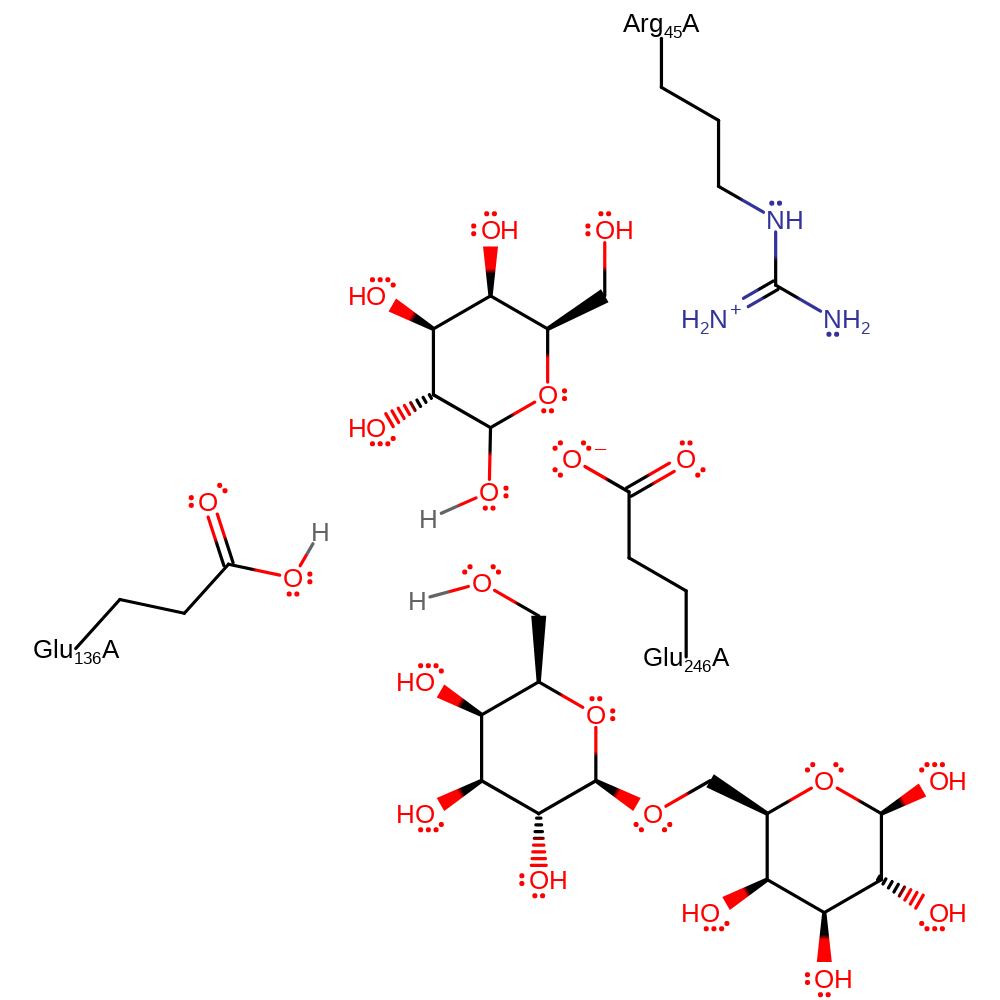 Download:
Download: