4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase
4-(cytidine 5'-diphospho0-2-C-methyl-D-erythritol (CDP-ME) kinase catalyses phosphorylation of the 2-hydroxyl group of CDP-ME, the fourth step of the dimethylallyl diphosphate (DMAPP) and isopentenyl diphosphate (IPP) biosynthesis pathways. IPP and DMAPP are universal precursors of isoprenoids, an important family of natural products, such as sterols, dilichols, triterpernes and ubiquinones and thus contribute to many biological functions. Two biosynthetic routes to IPP and DMAPP have evolved. Eukaryotes, archaebacteria and a few eubacteria uses the mevalonate pathway but in most eubacteria, algae, chloroplasts and cyanobacteria, the 1-deoxy-D-xylulose-5-phosphate (DOXP) pathway is used. The bacteria using DOXP pathway include some that cause serious human diseases and since the enzymes in DOXP pathway have no ortholog in human, they are the target for structure-based antimicrobial drug development.
Reference Protein and Structure
- Sequence
-
Q8FI04
 (2.7.1.148)
(2.7.1.148)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli CFT073 (Bacteria)

- PDB
-
1oj4
- Ternary complex of 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase
(2.01 Å)



- Catalytic CATH Domains
-
3.30.230.10
 (see all for 1oj4)
(see all for 1oj4)
- Cofactors
- Magnesium(2+) (1)
Enzyme Reaction (EC:2.7.1.148)
Enzyme Mechanism
Introduction
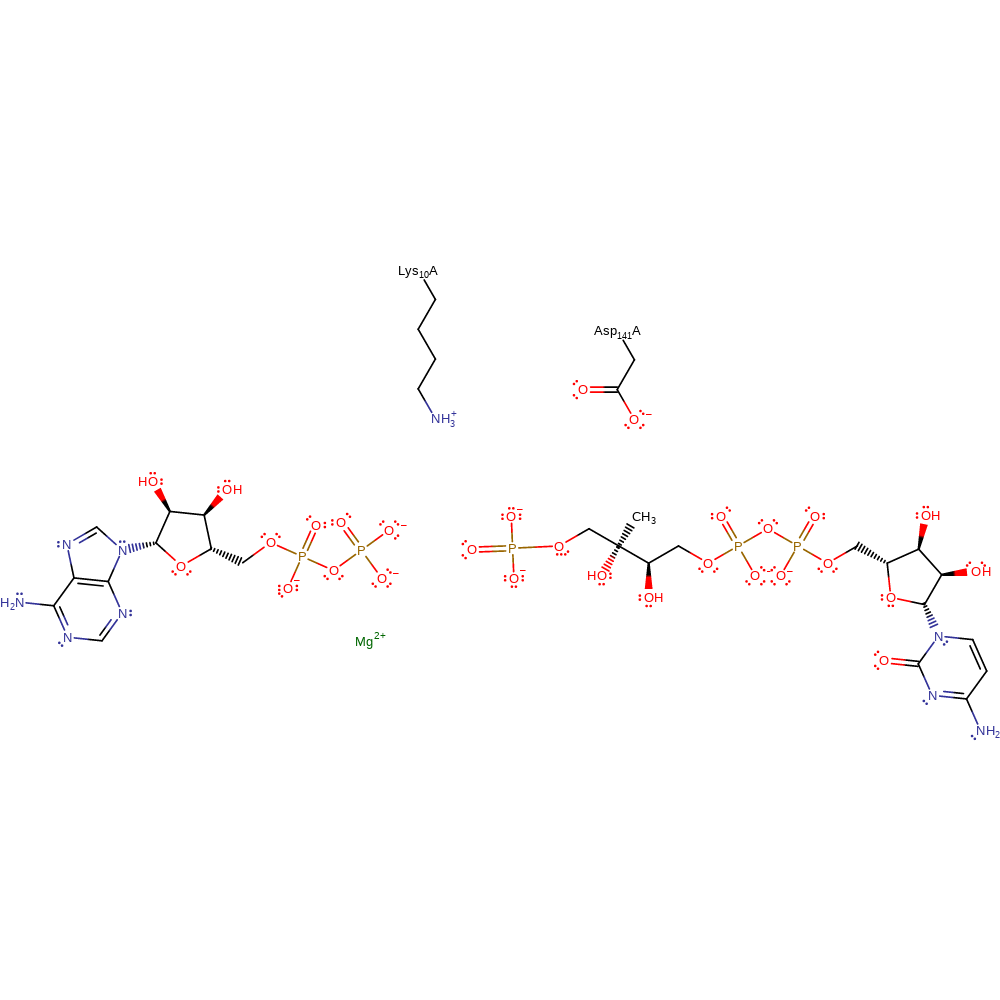
The catalysis is most likely to follow an associative in-line mechanism to form a pentacovalent intermediate. Asp141 acts as a base to deprotonate the hydroxyl group of CDP-ME to promote its nucleophilic attack on the terminal phosphate of ATP. Lys10 stabilises the transition state by interacting with the oxygen of the hydroxyl group. Additionally, a magnesium cation is needed for catalysis but no crystal structure of CDP-ME has been solved with the cofactor present.
Catalytic Residues Roles
| UniProt | PDB* (1oj4) | ||
| Lys10 | Lys10A | It stabilises the transition state by interacting with the oxygen atom of the substrate hydroxyl group. | hydrogen bond donor, electrostatic stabiliser |
| Asp141 | Asp141A | It acts as a base to deprotonate the hydroxyl group of CDP-ME to promote its nucleophilic attack on the terminal phosphate of ATP. |
activator, proton acceptor, proton donor |
Chemical Components
proton transfer, bimolecular nucleophilic substitution, overall product formed, overall reactant used, native state of enzyme regeneratedReferences
- Miallau L et al. (2003), Proc Natl Acad Sci U S A, 100, 9173-9178. Biosynthesis of isoprenoids: Crystal structure of 4-diphosphocytidyl-2C-methyl-D-erythritol kinase. DOI:10.1073/pnas.1533425100. PMID:12878729.
- Frank A et al. (2017), Chem Rev, 117, 5675-5703. The Methylerythritol Phosphate Pathway to Isoprenoids. DOI:10.1021/acs.chemrev.6b00537. PMID:27995802.
- Wada T et al. (2003), J Biol Chem, 278, 30022-30027. Crystal Structure of 4-(Cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase, an Enzyme in the Non-mevalonate Pathway of Isoprenoid Synthesis. DOI:10.1074/jbc.m304339200. PMID:12771135.

Step 1. Asp141 deprotonates the hydroxyl group on the substrate. This increases the nucleophilicity of the substract to then attack the gamma phosphate on ATP in an inline nucleophilic substitution.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys10A | electrostatic stabiliser |
| Asp141A | activator |
| Lys10A | hydrogen bond donor |
| Asp141A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, overall product formed, overall reactant usedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp141A | proton donor |






 Download:
Download: