Kanamycin nucleotidyltransferase
Kanamycin nucleotidyltransferase (KNTase) is a plasmid encoded enzyme responsible for some types of bacterial resistance to aminoglycosides. It catalyses the transfer of a nucleoside monophosphate group onto the 4' hydroxyl group of kanamycin, causing deactivation of this drug. The enzyme is also capable of deactivating many other drugs including neomycins, and can utilise ATP, GTP, CTP, TTP or UTP as the NMP donor.
Reference Protein and Structure
- Sequence
-
P05057
 (2.7.7.-)
(2.7.7.-)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Staphylococcus aureus (Bacteria)

- PDB
-
1kny
- KANAMYCIN NUCLEOTIDYLTRANSFERASE
(2.5 Å)



- Catalytic CATH Domains
-
3.30.460.10
 1.20.120.330
1.20.120.330  (see all for 1kny)
(see all for 1kny)
- Cofactors
- Magnesium(2+) (1)
Enzyme Mechanism
Introduction
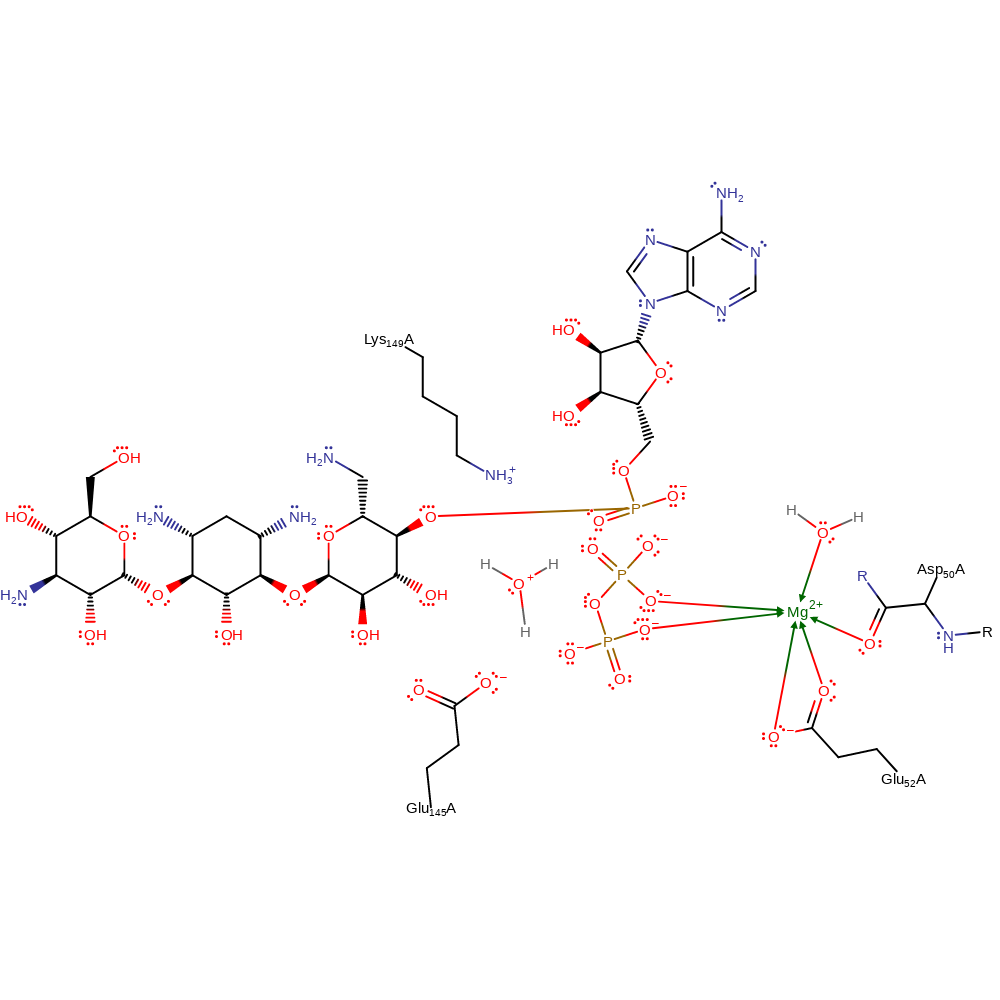
Kanamycin nucleotidyltransferase catalyses a single in-line displacement attack by the 4' hydroxyl group of kanamycin on the alpha phosphate of the NTP. The leaving group is pyrophosphate. As is typical for enzymatic phosphodiester reactions, the substitution is concerted with a slightly associative transition-state. Glu 145 acts as a base to deprotonate the attacking 4' hydroxyl group, while Lys 149 functions to stabilise the accumulation of negative charge on the alpha phosphate oxygens in the transition state. An Mg2+ ion coordinates the beta and gamma phosphates, so stabilising the departing pyrophosphate leaving group. Deprotonation of Glu 145 can occur to regenerate the active site ready for another round of catalysis such as a water molecule picking the proton up.
Catalytic Residues Roles
| UniProt | PDB* (1kny) | ||
| Glu52, Asp50 (main-C) | Glu52B, Asp50B (main-C) | Proposed to coordinate to a magnesium ion which in turn stabilises the pyrophosphate leaving group. | metal ligand |
| Glu145 | Glu145A | Acts as a general base to deprotonate the 4' hydroxyl group of the antibiotic as it attacks the alpha phosphate of the NTP. | activator, proton acceptor, proton donor |
| Lys149 | Lys149A | Provides a positive charge to stabilise accumulation of negative charge on the alpha phosphate oxygens in the transition state. | electrostatic stabiliser, polar interaction |
Chemical Components
proton transfer, bimolecular nucleophilic substitution, overall reactant used, overall product formed, native state of enzyme regeneratedReferences
- Gerratana B et al. (2001), Biochemistry, 40, 2972-2977. Characterization of the Transition-State Structure of the Reaction of Kanamycin Nucleotidyltransferase by Heavy-Atom Kinetic Isotope Effects†. DOI:10.1021/bi002557x. PMID:11258909.
- Cleland WW et al. (2006), Chem Rev, 106, 3252-3278. Enzymatic Mechanisms of Phosphate and Sulfate Transfer. DOI:10.1021/cr050287o. PMID:16895327.
- Chen-Goodspeed M et al. (1999), Bioorg Chem, 27, 395-408. Kinetic Mechanism of Kanamycin Nucleotidyltransferase from Staphylococcus aureus. DOI:10.1006/bioo.1999.1144.
- Pedersen LC et al. (1995), Biochemistry, 34, 13305-13311. Structural investigation of the antibiotic and ATP-binding sites in kanamycin nucleotidyltransferase. DOI:10.1021/bi00041a005. PMID:7577914.

Step 1. Glu45 abstracts the proton on the 4' hydroxyl group in kanamycin. This activates the group for nucleophilic attack of the alpha phosphate on ATP. Lys149 stabilises the negatively charged transition state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys149A | electrostatic stabiliser |
| Glu52B | metal ligand |
| Asp50B (main-C) | metal ligand |
| Glu145A | activator |
| Lys149A | polar interaction |
| Glu145A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, overall reactant used, overall product formed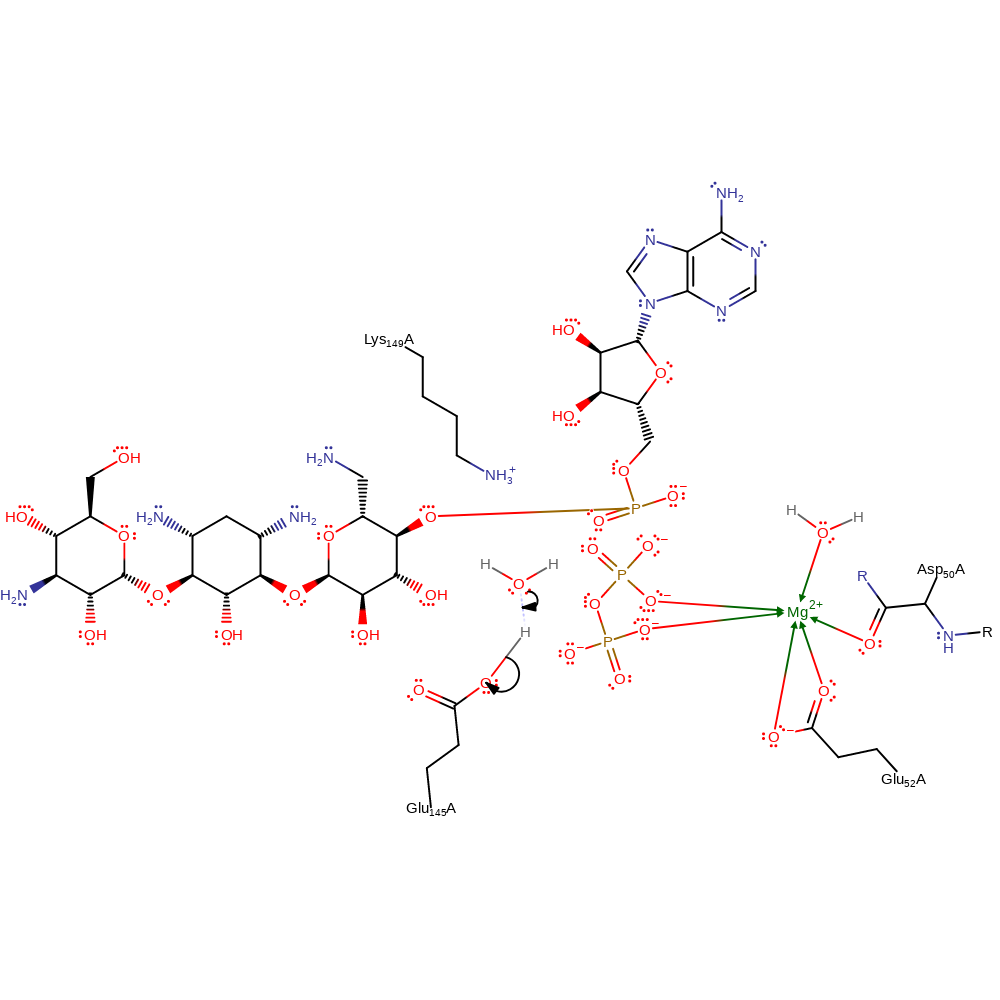
Step 2. Solvent water molecule deprotonates Glu145 to regenerate active site.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu145A | proton donor |




 Download:
Download: