Signal peptidase I
Escherichia coli type I signal peptidase is a membrane-bound serine endopeptidase that cleaves the amino-terminal signal sequence from secretory proteins and some membrane proteins. Evolutionarily the enzyme belongs to the protease clan SF and the protease family S26.
Reference Protein and Structure
- Sequence
-
P00803
 (3.4.21.89)
(3.4.21.89)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1t7d
- Crystal structure of Escherichia coli type I signal peptidase in complex with a lipopeptide inhibitor
(2.47 Å)



- Catalytic CATH Domains
-
2.10.109.10
 (see all for 1t7d)
(see all for 1t7d)
Enzyme Reaction (EC:3.4.21.89)
Enzyme Mechanism
Introduction
Signal peptidase uses a Ser-Lys dyad to catalyse attack on the si-face of the peptide bond. Ser 91 acts as a nucleophile to attack the carbonyl group while Lys 146 deprotonates the attacking Ser 91. The resulting tetrahedral intermediate is stabilised by an oxyanion hole consisting of the backbone NH of Ser 91 and the side chain hydroxyl of Ser 89. Collapse of the tetrahedral intermediate with protonation of the departing amine by Lys 146 generates an acyl-enzyme intermediate; this is then hydrolysed by a water molecule that is deprotonated by Lys 146. Lys 146 is located in a hydrophobic environment in the protein; this reduces its pKa and so allows it to carry out its role as an acid/base.
Catalytic Residues Roles
| UniProt | PDB* (1t7d) | ||
| Ser279 | Ser278(205)B | Stabilises the Lys residue. | electrostatic stabiliser |
| Ser89 | Ser88(15)B | Side chain hydroxyl forms part of the oxyanion hole that stabilises the tetrahedral intermediate. | electrostatic stabiliser |
| Ser91 (main-N), Ser91 | Ser90(17)B (main-N), Ser90(17)B | Attacks the peptide bond to form an acyl enzyme intermediate. Backbone NH forms part of the oxyanion hole that stabilises the tetrahedral intermediate. | electrostatic stabiliser |
| Lys146 | Lys145(72)B | Deprotonates Ser 91 as Ser 91 attacks the peptide carbonyl. Protonates the departing amine group during collapse of the tetrahedral intermediate. Deprotonates a water molecule during hydrolysis of the acyl-enzyme intermediate. | proton acceptor, proton donor |
Chemical Components
proton transfer, bimolecular nucleophilic addition, intermediate formation, overall reactant used, rate-determining step, unimolecular elimination by the conjugate base, intermediate collapse, overall product formed, native state of enzyme regeneratedReferences
- Paetzel M et al. (2004), J Biol Chem, 279, 30781-30790. Crystallographic and Biophysical Analysis of a Bacterial Signal Peptidase in Complex with a Lipopeptide-based Inhibitor. DOI:10.1074/jbc.m401686200. PMID:15136583.
- Paetzel M (2014), Biochim Biophys Acta, 1843, 1497-1508. Structure and mechanism of Escherichia coli type I signal peptidase. DOI:10.1016/j.bbamcr.2013.12.003. PMID:24333859.
- Carlos JL et al. (2000), Biochemistry, 39, 7276-7283. Mutational Evidence of Transition State Stabilization by Serine 88 inEscherichia coliType I Signal Peptidase†,‡. DOI:10.1021/bi000301l. PMID:10852727.
- Paetzel M et al. (1998), Nature, 396, 186-190. Crystal structure of a bacterial signal peptidase in complex with a beta-lactam inhibitor. DOI:10.1038/24196. PMID:9823901.
- Tschantz WR et al. (1993), J Biol Chem, 268, 27349-27354. A serine and a lysine residue implicated in the catalytic mechanism of the Escherichia coli leader peptidase. PMID:8262975.
- Sung M et al. (1992), J Biol Chem, 267, 13154-13159. Identification of potential active-site residues in the Escherichia coli leader peptidase. PMID:1618816.
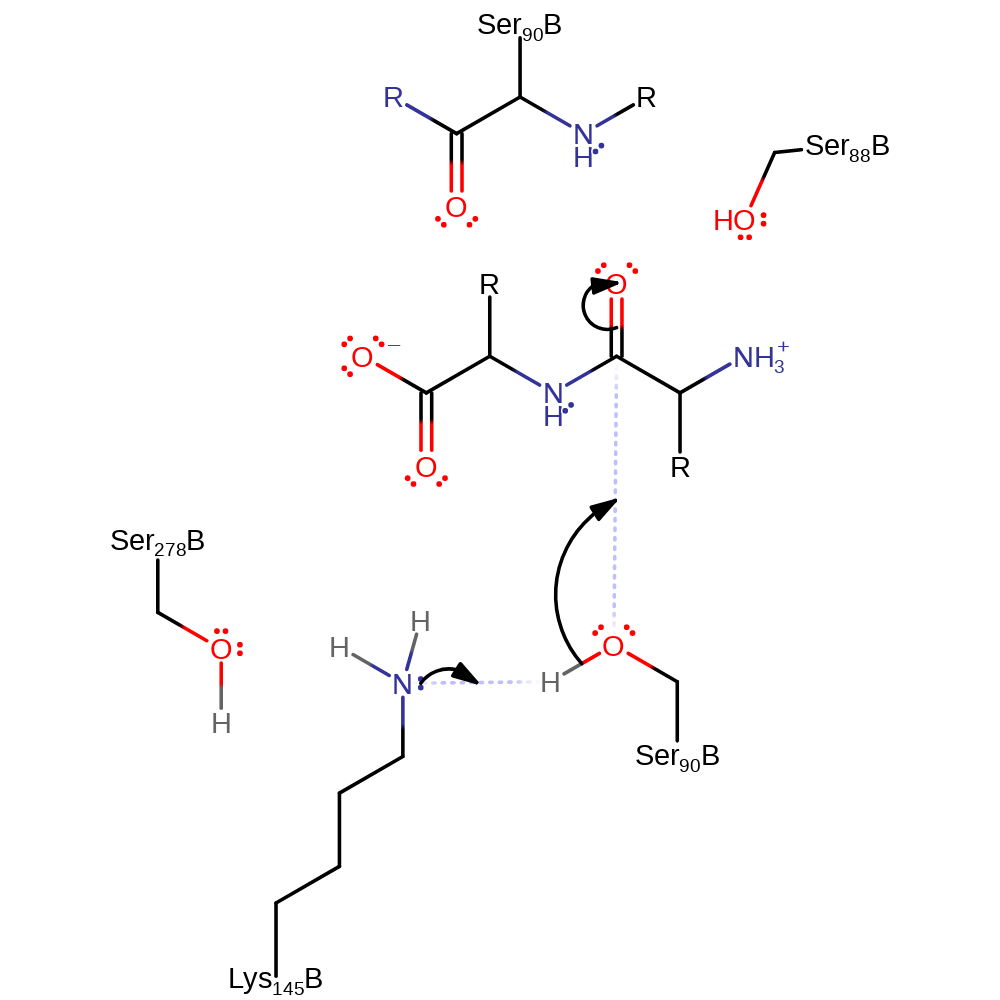
Step 1. Lys 146 deprotonates Ser 91 which activates it to nucleophilically attack the carbon of the carbonyl to form the oxyanion intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser88(15)B | electrostatic stabiliser |
| Ser90(17)B (main-N) | electrostatic stabiliser |
| Ser278(205)B | electrostatic stabiliser |
| Ser90(17)B | proton donor |
| Lys145(72)B | proton acceptor |
| Ser90(17)B | nucleophile |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used, rate-determining step
Step 2. Lys 146 protonates the amine of the peptide bond which results in the collapse of the tetrahedral intermediate and the cleavage of the peptide bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser88(15)B | electrostatic stabiliser |
| Ser90(17)B (main-N) | electrostatic stabiliser |
| Ser278(205)B | electrostatic stabiliser |
| Lys145(72)B | proton donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transfer, intermediate collapse, overall product formed
Step 3. Lys 146 abstracts a proton from a water which activates it to attack the carbon of the ester bond in a nucleophilic addition once again producing the oxyanion intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser88(15)B | electrostatic stabiliser |
| Ser90(17)B (main-N) | electrostatic stabiliser |
| Ser278(205)B | electrostatic stabiliser |
| Lys145(72)B | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used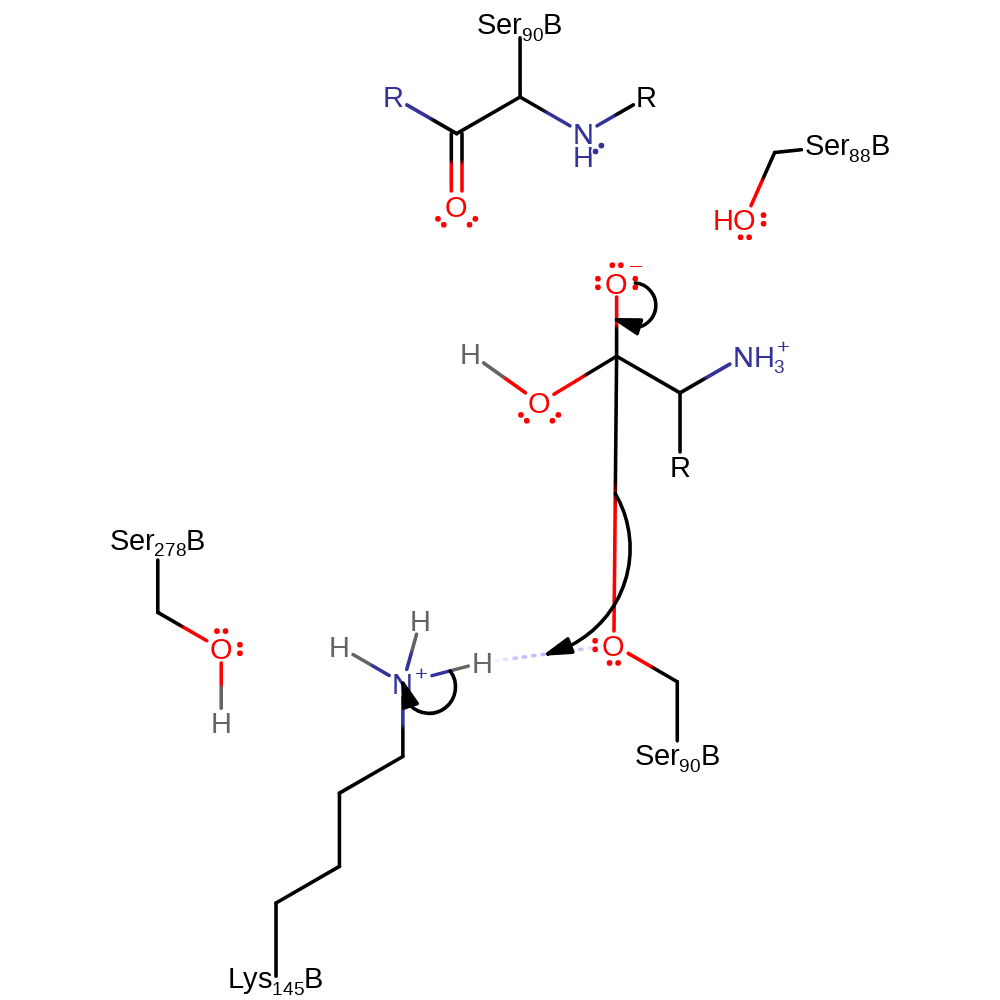
Step 4. The tetrahedral intermediate collapse again resulting in the release of Ser91 which then accepts a proton from Lys 146 returning the active site to its native state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser88(15)B | electrostatic stabiliser |
| Ser90(17)B (main-N) | electrostatic stabiliser |
| Ser278(205)B | electrostatic stabiliser |
| Lys145(72)B | proton donor |
| Ser90(17)B | nucleofuge, proton acceptor |



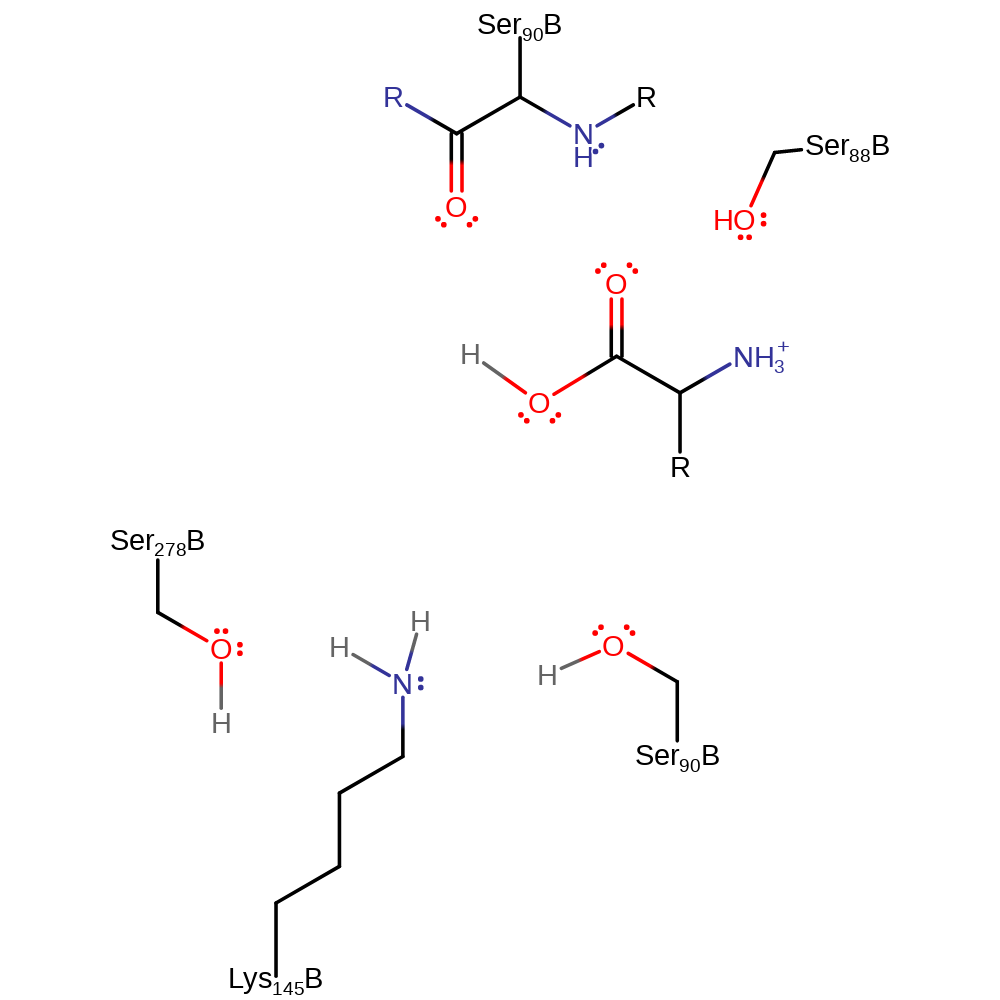 Download:
Download: