Methylaspartate mutase
This vitamin B12 dependent enzyme catalyses the carbon skeleton rearrangement of L-glutamate to L-threo-3-methylaspartate ((2S,3S)-3-methylaspartate). It is involved in the subpathway that synthesises acetate and pyruvate from L-glutamate.
Reference Protein and Structure
- Sequences
-
P80077
 (5.4.99.1)
(5.4.99.1)
P80078 (5.4.99.1)
(5.4.99.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Clostridium cochlearium (Bacteria)

- PDB
-
1cb7
- GLUTAMATE MUTASE FROM CLOSTRIDIUM COCHLEARIUM RECONSTITUTED WITH METHYL-COBALAMIN
(2.0 Å)



- Catalytic CATH Domains
-
3.40.50.280
 3.20.20.240
3.20.20.240  (see all for 1cb7)
(see all for 1cb7)
- Cofactors
- Methylcobalamin (1) Metal MACiE
Enzyme Reaction (EC:5.4.99.1)
Enzyme Mechanism
Introduction
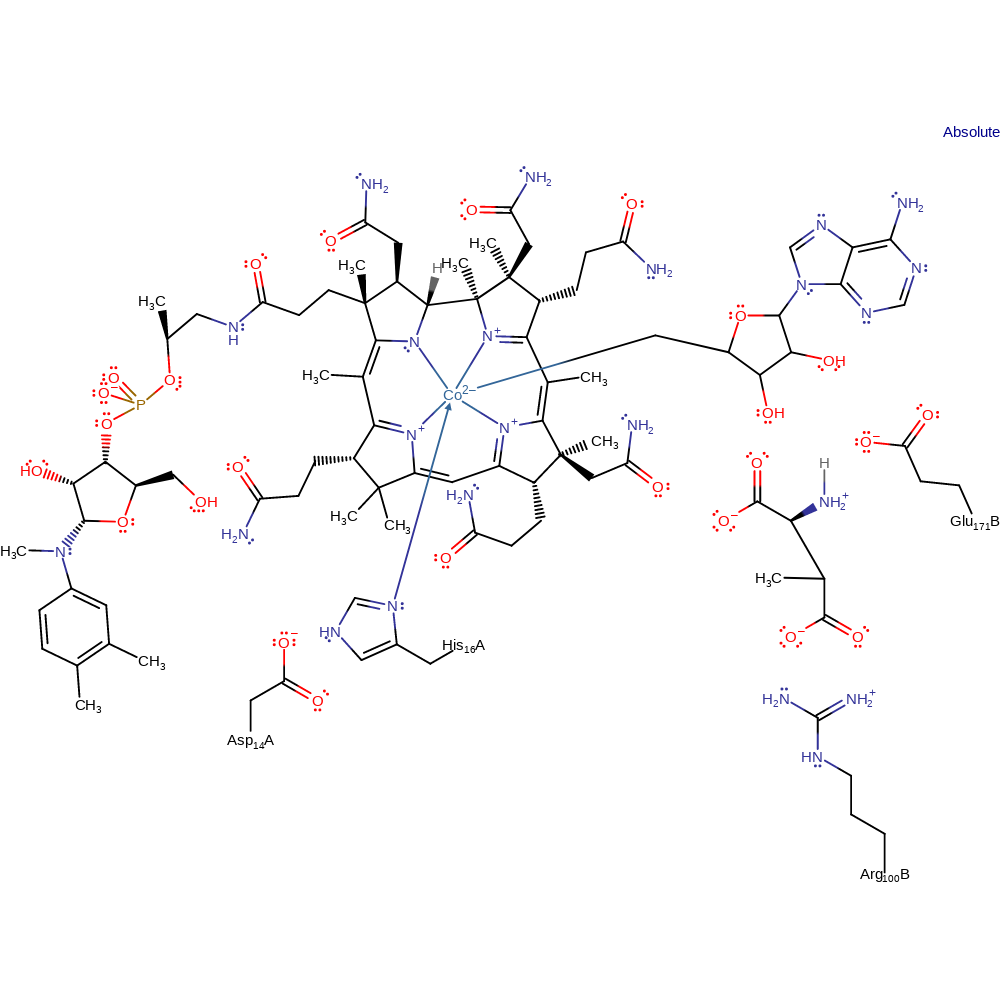
Glutamate mutase (Glm) equilibrates (S)-glutamate with (2S,3S)-3-methylaspartate.
The Co-C bond of AdoCbl cleaves to produce a 5'-deoxyadenosyl (Ado) radical and Cbl(II). This is coupled to the abstraction of the pro-S hydrogen on C4 of glutamate by the ado radical, which occurs by quantum tunnelling of the hydrogen. The glutamate radical fragments to form the glycyl radical and acrylic acid. These two species recombine to form the methylaspartate radical, which then abstracts a proton from Ado-H to form methylaspartate and the Ado radical. The radical recombines with Cbl(II) to form the resting cofactor.
Catalytic Residues Roles
| UniProt | PDB* (1cb7) | ||
| His16 | His16A | Forms the axial ligand to the cobalt ion of the cobalamin cofactor. It is thought to stabilise Cbl(II), which will lower the activation barrier for homolysis. | metal ligand, electrostatic stabiliser |
| Glu171 | Glu171B | Acts as a general acid/base. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, electrostatic stabiliser |
| Arg100 | Arg100B | Involved in stabilising and binding the substrate. | electrostatic stabiliser |
| Asp14 | Asp14A | Asp14 forms a charged hydrogen bond with His16. It is thought to affect the strength of the coordination of His16 to the cobalt of the cofactor, thereby influencing the stabilisation of Cbl(II). | electrostatic stabiliser |
Chemical Components
radical formation, homolysis, cofactor used, coordination to a metal ion, decoordination from a metal ion, intermediate formation, hydrogen transfer, radical propagation, overall reactant used, unimolecular homolytic elimination, proton transfer, intermediate collapse, bimolecular homolytic addition, overall product formed, intermediate terminated, colligation, radical termination, native state of enzyme regenerated, native state of cofactor regeneratedReferences
- Madhavapeddi P et al. (2001), Chem Biol, 8, 1143-1149. The role of the active site glutamate in the rearrangement of glutamate to 3-methylaspartate catalyzed by adenosylcobalamin-dependent glutamate mutase. DOI:10.1016/s1074-5521(01)00081-3. PMID:11755393.
- Rommel JB et al. (2012), J Phys Chem B, 116, 13682-13689. Role of Tunneling in the Enzyme Glutamate Mutase. DOI:10.1021/jp308526t. PMID:23127187.
- Sandala GM et al. (2007), J Am Chem Soc, 129, 1623-1633. Toward an Improved Understanding of the Glutamate Mutase System. DOI:10.1021/ja066432c. PMID:17249667.
- Yoon M et al. (2006), Biochemistry, 45, 11650-11657. Reaction of Adenosylcobalamin-Dependent Glutamate Mutase with 2-Thiolglutarate†. DOI:10.1021/bi061067n. PMID:16981724.
- Brooks AJ et al. (2005), Biochemistry, 44, 15167-15181. Electronic Structure Studies of the Adenosylcobalamin Cofactor in Glutamate Mutase†. DOI:10.1021/bi051094y. PMID:16285720.
- Cheng MC et al. (2005), Biochemistry, 44, 2686-2691. Isotope Effects for Deuterium Transfer between Substrate and Coenzyme in Adenosylcobalamin-Dependent Glutamate Mutase†. DOI:10.1021/bi047662b. PMID:15709782.
- Xia L et al. (2004), Biochemistry, 43, 3238-3245. Role of Arg100 in the Active Site of Adenosylcobalamin-Dependent Glutamate Mutase†. DOI:10.1021/bi0357558. PMID:15023074.
- Banerjee R (2003), Chem Rev, 103, 2083-2094. Radical Carbon Skeleton Rearrangements: Catalysis by Coenzyme B12-Dependent Mutases. DOI:10.1021/cr0204395. PMID:12797824.
- Gruber K et al. (2002), Curr Opin Chem Biol, 6, 598-603. Coenzyme B12 dependent glutamate mutase. DOI:10.1016/s1367-5931(02)00368-x.
- Marsh EN et al. (2001), Curr Opin Chem Biol, 5, 499-505. Adenosylcobalamin-dependent isomerases: new insights into structure and mechanism. DOI:10.1016/s1367-5931(00)00238-6. PMID:11578922.
- Wetmore SD et al. (2001), J Am Chem Soc, 123, 7963-7972. Interconversion of (S)-Glutamate and (2S,3S)-3-Methylaspartate: A Distinctive B12-Dependent Carbon-Skeleton Rearrangement. DOI:10.1021/ja004246f. PMID:11506551.
- Gruber K et al. (2001), Angew Chem Int Ed Engl, 40, 3377-3380. Radical Shuttling in a Protein: Ribose Pseudorotation Controls Alkyl-Radical Transfer in the Coenzyme B12 Dependent Enzyme Glutamate Mutase. DOI:10.1002/1521-3773(20010917)40:18<3377::aid-anie3377>3.0.co;2-8. PMID:11592143.
- Huhta MS et al. (2001), Biochem J, 355, 131-137. Protein-coenzyme interactions in adenosylcobalamin-dependent glutamate mutase. PMID:11256957.
- Roymoulik I et al. (2000), Biochemistry, 39, 10340-10346. Rearrangement ofl-2-Hydroxyglutarate tol-threo-3-Methylmalate Catalyzed by Adenosylcobalamin-Dependent Glutamate Mutase†. DOI:10.1021/bi000121b.
- Marsh EN (2000), Bioorg Chem, 28, 176-189. Review Article Coenzyme-B12-Dependent Glutamate Mutase. DOI:10.1006/bioo.2000.1168. PMID:10915555.
- Reitzer R et al. (1999), Structure, 7, 891-902. Glutamate mutase from Clostridium cochlearium: the structure of a coenzyme B12-dependent enzyme provides new mechanistic insights. DOI:10.1016/s0969-2126(99)80116-6. PMID:10467146.
- Chih H et al. (1999), Biochemistry, 38, 13684-13691. Pre-Steady-State Kinetic Investigation of Intermediates in the Reaction Catalyzed by Adenosylcobalamin-Dependent Glutamate Mutase†. DOI:10.1021/bi991064t.
- Marsh EN et al. (1998), Biochemistry, 37, 11864-11872. Coupling of Cobalt−Carbon Bond Homolysis and Hydrogen Atom Abstraction in Adenosylcobalamin-Dependent Glutamate Mutase†. DOI:10.1021/bi980512e. PMID:9718309.
- Bothe H et al. (1998), Biochemistry, 37, 4105-4113. Identification of the 4-Glutamyl Radical as an Intermediate in the Carbon Skeleton Rearrangement Catalyzed by Coenzyme B12-Dependent Glutamate Mutase fromClostridiumcochlearium†. DOI:10.1021/bi971393q. PMID:9521732.
- Tollinger M et al. (1998), Structure, 6, 1021-1033. How a protein prepares for B12 binding: structure and dynamics of the B12-binding subunit of glutamate mutase from Clostridium tetanomorphum. DOI:10.1016/s0969-2126(98)00103-8. PMID:9739092.
- Zelder O et al. (1995), FEBS Lett, 369, 252-254. Coordination of a histidine residue of the protein-component S to the cobalt atom in coenzyme B12-dependent glutamate mutase fromClostridium cochlearium. DOI:10.1016/0014-5793(95)00762-x. PMID:7649266.
- Zelder O et al. (1994), Eur J Biochem, 226, 577-585. Characterization of the coenzyme-B12-dependent glutamate mutase from Clostridium cochlearium produced in Escherichia coli. PMID:7880251.
- Leutbecher U et al. (1992), Eur J Biochem, 205, 759-765. Glutamate mutase from Clostridium cochlearium. Purification, cobamide content and stereospecific inhibitors. PMID:1315276.
Step 1. The Co(II)-C bond of the B12 cofactor undergoes homoloysis, forming the adenosyl radical and Co(I).
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp14A | electrostatic stabiliser |
| His16A | metal ligand |
| Arg100B | electrostatic stabiliser |
Chemical Components
radical formation, homolysis, cofactor used, coordination to a metal ion, decoordination from a metal ion, intermediate formationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp14A | electrostatic stabiliser |
| His16A | metal ligand, electrostatic stabiliser |
| Glu171B | hydrogen bond acceptor, electrostatic stabiliser |
| Arg100B | electrostatic stabiliser |
Chemical Components
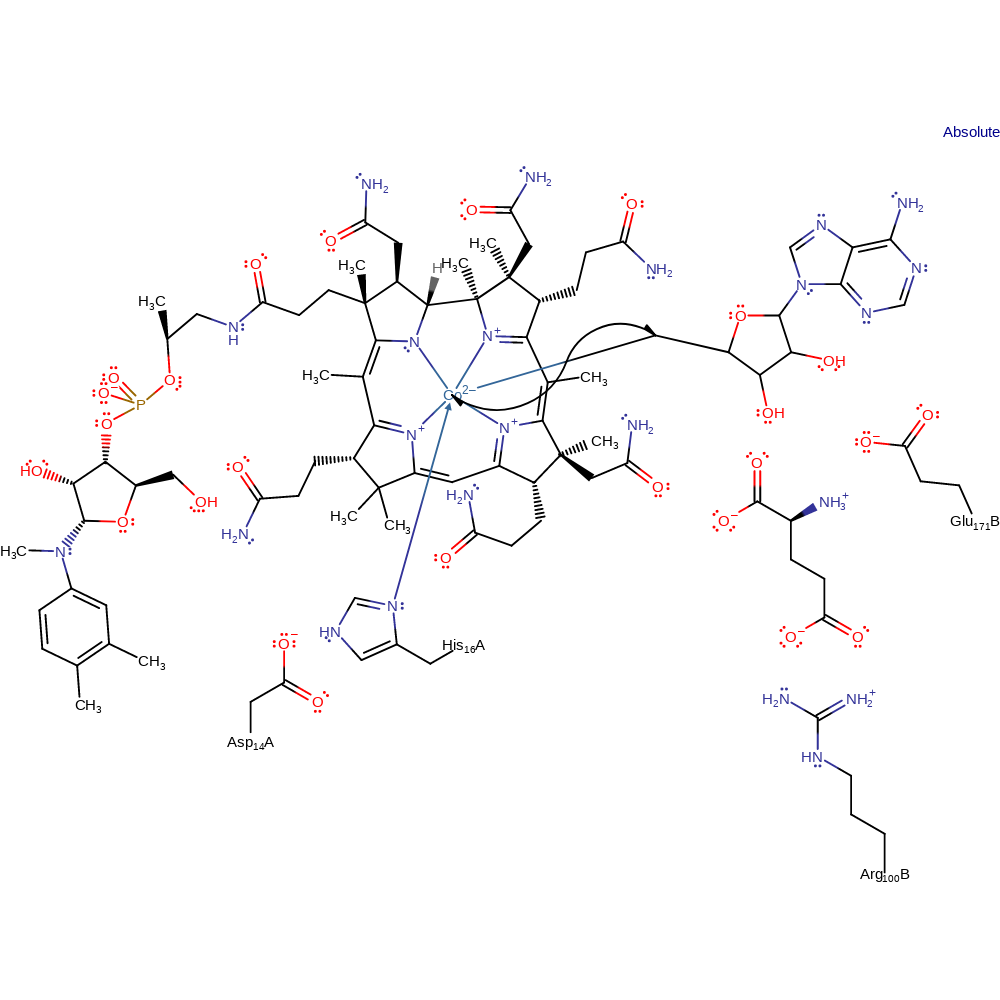
hydrogen transfer, radical propagation, overall reactant used, intermediate formation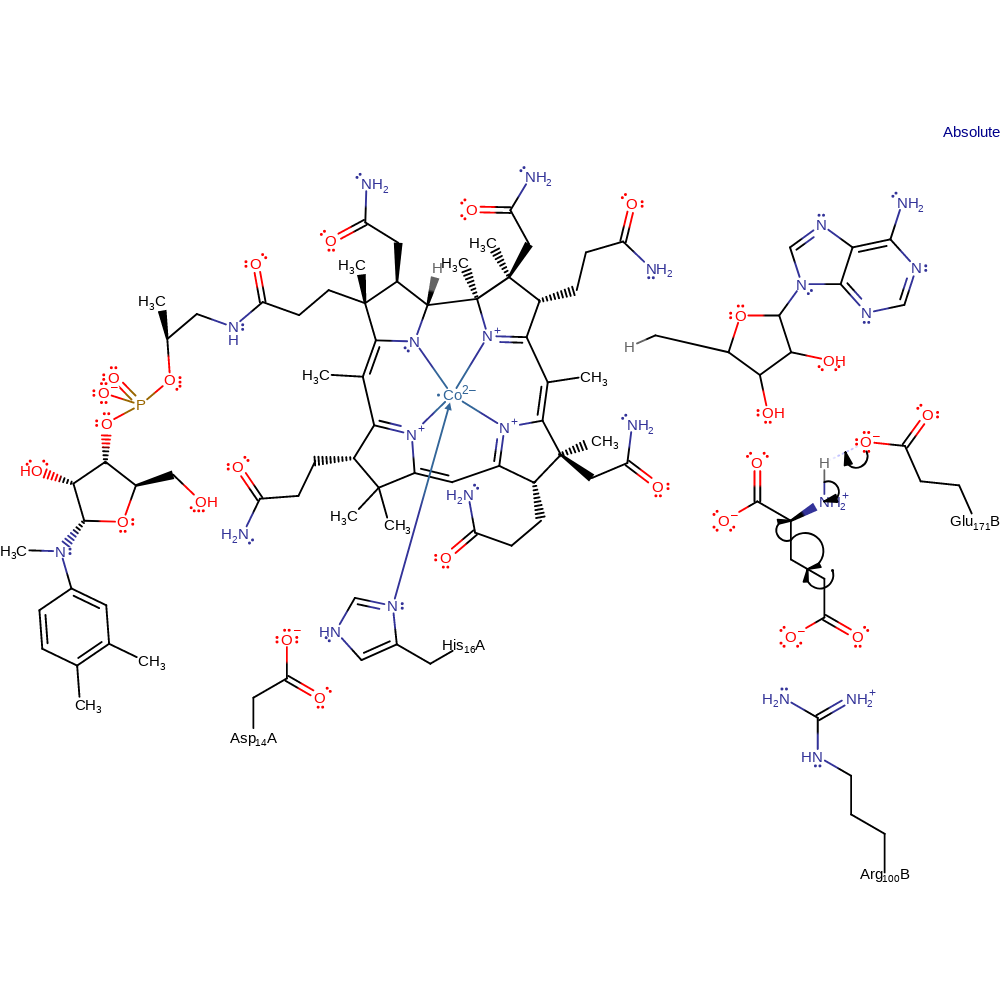
Step 3. The substrate undergoes homolytic elimination producing prop-2-enoic acid and a glycyl radical with concomitant deprotonation of the positively charged amine of the glycyl radical by Glu171B.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp14A | electrostatic stabiliser |
| Glu171B | hydrogen bond acceptor, electrostatic stabiliser |
| His16A | electrostatic stabiliser, metal ligand |
| Arg100B | electrostatic stabiliser |
| Glu171B | proton acceptor |
Chemical Components
ingold: unimolecular homolytic elimination, radical propagation, proton transfer, intermediate formation, intermediate collapse
Step 4. The glycyl radical adds to the double bond of prop-2-enoic acid, forming the product radical with concomitant deprotonation of Glu171B by the glycyl amine.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp14A | electrostatic stabiliser |
| Glu171B | hydrogen bond donor |
| His16A | electrostatic stabiliser, metal ligand |
| Arg100B | electrostatic stabiliser |
| Glu171B | proton donor |
Chemical Components
radical propagation, ingold: bimolecular homolytic addition, proton transfer, intermediate formation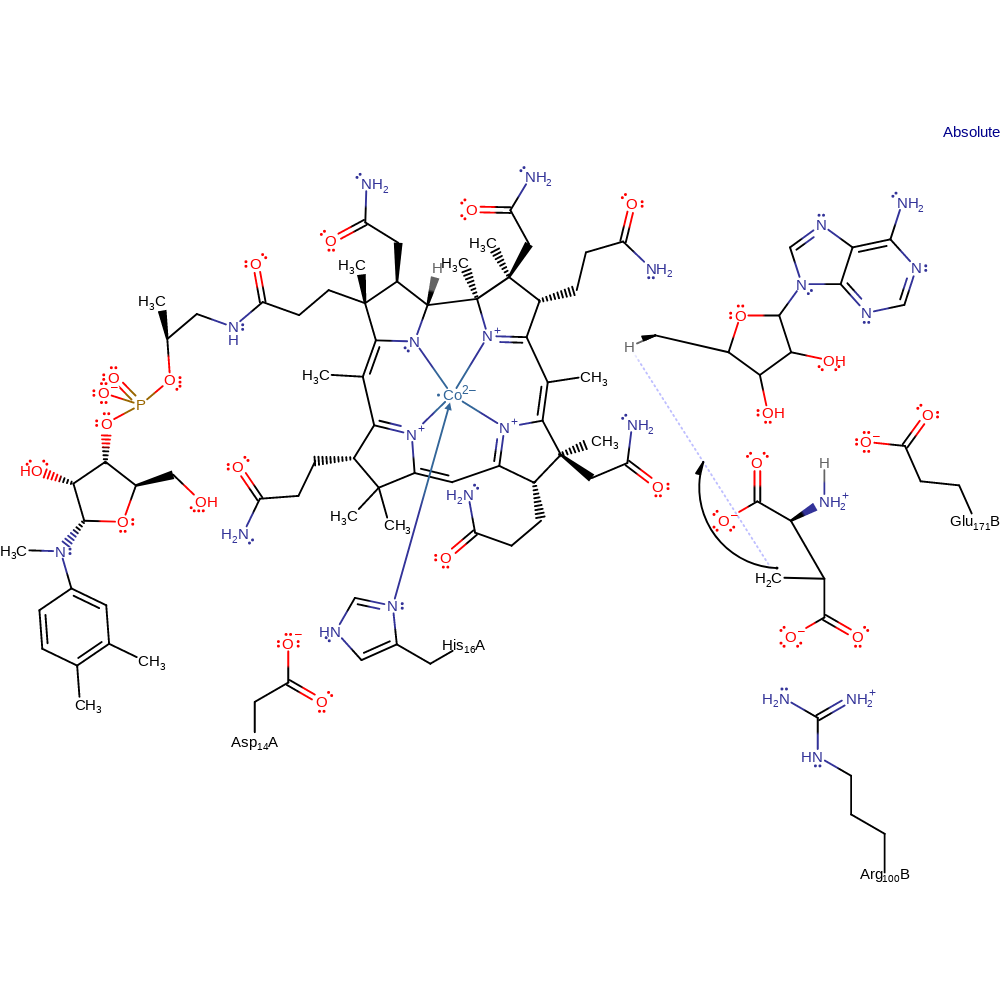
Step 5. The substrate radical abstracts a hydrogen from the adenosine, re-forming the adenosyl radical.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp14A | electrostatic stabiliser |
| Glu171B | hydrogen bond acceptor, electrostatic stabiliser |
| His16A | electrostatic stabiliser, metal ligand |
| Arg100B | electrostatic stabiliser |
Chemical Components
radical propagation, hydrogen transfer, overall product formed, intermediate terminated, intermediate formation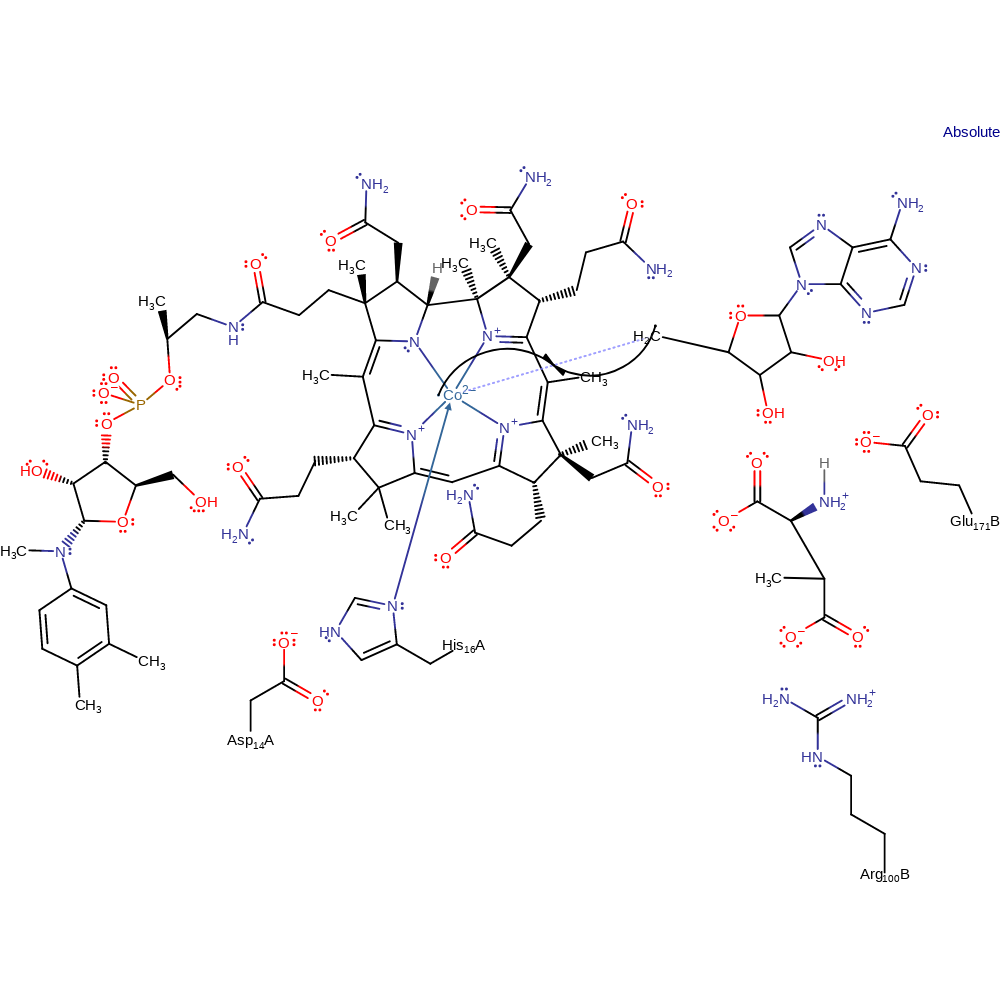
Step 6. The adenosyl radical and Co(I) undergo colligation, reforming the B12 cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp14A | electrostatic stabiliser |
| His16A | metal ligand, electrostatic stabiliser |



 Download:
Download: