Opine dehydrogenase
N-(1-D-carboxylethyl)-L-norvaline dehydrogenase is part of the (D,L) stereochemistry family of opine dehydrogenases and is able to catalyse the reaction of pyruvate with hydrophobic L amino acids to form opines through reductive condensation using NAD(P)H as a cofactor. Of particular interest is the stereochemical specificity of this enzyme as its reaction is a key step in a number of synthetic routes for pharmaceuticals. The enzyme displays some sequence homology with (L,L) opine dehydrogenases and both families display homology with 2-hydroxy acid dehydrogenases.
Reference Protein and Structure
- Sequence
-
Q44297
 (1.5.1.28)
(1.5.1.28)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Arthrobacter sp. 1C (Bacteria)

- PDB
-
1bg6
- CRYSTAL STRUCTURE OF THE N-(1-D-CARBOXYLETHYL)-L-NORVALINE DEHYDROGENASE FROM ARTHROBACTER SP. STRAIN 1C
(1.8 Å)



- Catalytic CATH Domains
-
1.10.1040.10
 (see all for 1bg6)
(see all for 1bg6)
Enzyme Reaction (EC:1.5.1.28)
Enzyme Mechanism
Introduction
The mechanism proposal is based on the similarity in the chemistry of the opine dehydrogenase family to that of the amino acid dehydrogenases. The reaction is believed to proceed via nucleophilic attack by the L amino acid on the pyruvate with a histidine residue acting as a general acid, allowing the formation of a carbinolamine intermediate. Dehydration of this intermediate, again facilitated by acid-base action of Histidine, results in an imine intermediate which is then reduced by hydride transfer from NAD(P)H to give the product.
Catalytic Residues Roles
| UniProt | PDB* (1bg6) | ||
| Asp297 | Asp297A | Acts to alter the pKa of His 202 to allow it to function as a general acid-base catalyst at physiological pH and thus catalyse the reaction efficiently. Thus is part of an Asp-His proton relay system. | electrostatic stabiliser |
| His202 | His202A | Protonates the carbonyl oxygen resulting in a carbinolamine intermediate which proceeds via hydride transfer from the cofactor to form the product. | proton acceptor, proton donor |
Chemical Components
proton transfer, bimolecular nucleophilic addition, overall reactant used, intermediate formation, dehydration, aromatic unimolecular elimination by the conjugate base, hydride transfer, overall product formed, intermediate collapseReferences
- Britton KL et al. (1998), Nat Struct Biol, 5, 593-601. Crystal structure and active site location of N-(1-D-carboxylethyl)-L-norvaline dehydrogenase. DOI:10.1038/854. PMID:9665174.

Step 1. Catalysis begins with nucleophilic attack of the amino group of L-2-aminopentanoic acid on the carbonyl of pyruvate, forming a carbinolamine intermediate. His202 acts as an acid and donates a proton to the developing alkoxide on pyruvate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp297A | electrostatic stabiliser |
| His202A | proton donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, overall reactant used, intermediate formation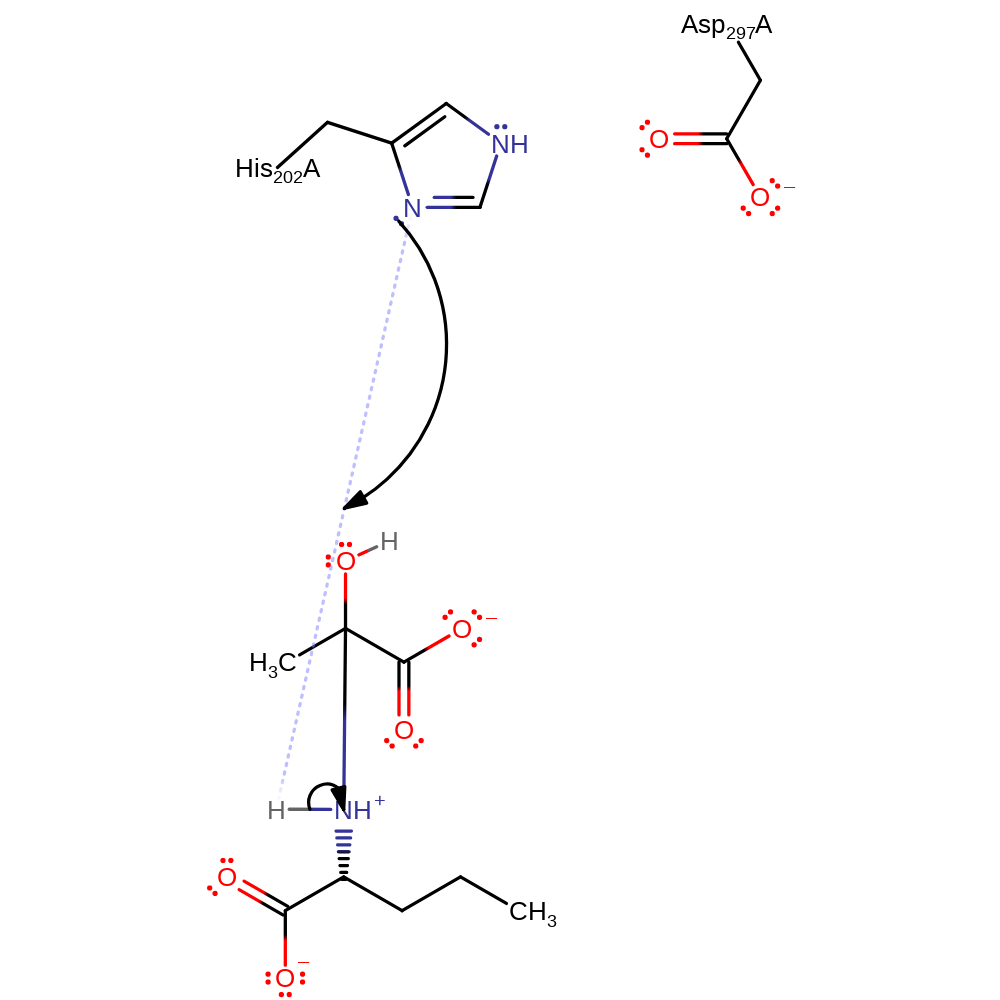
Step 2. His202 acts as a base to deprotonate the amine group of the intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp297A | electrostatic stabiliser |
| His202A | proton acceptor |
Chemical Components
proton transfer
Step 3. The hydroxyl group accepts a proton from His202 and the nitrogen lone pair facilitates dehydration of the intermediate, forming and imine intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His202A | proton donor |
Chemical Components
dehydration, proton transfer
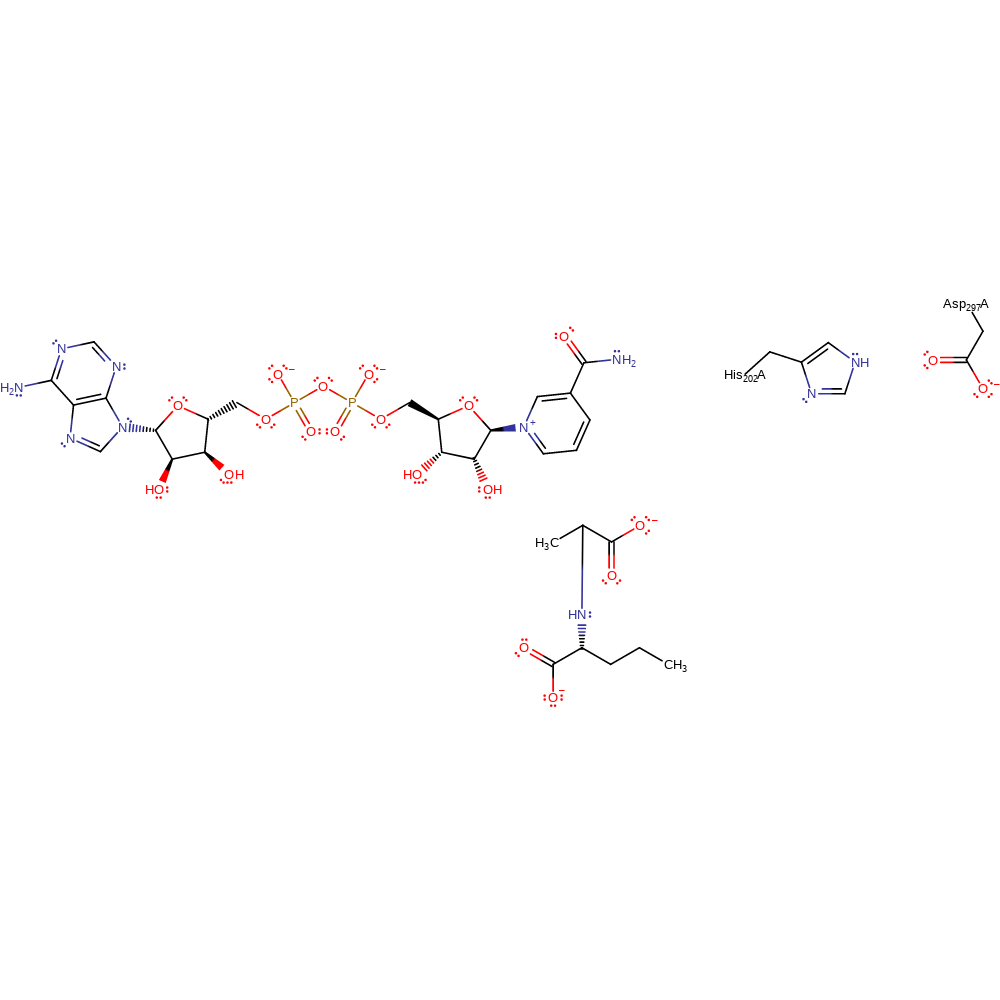
Step 4. Hydride transfer from NADH to the imine intermediate occurs, forming the product and NAD.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|







 Download:
Download: