Glutamate dehydrogenase
Glutamate dehydrogenases catalyse the reversible oxidative deamination of L-glutamate, with NAD(P)H as cofactor. Glutamatc dehydrogcnases arc widely distributed throughout the cukaryotic, cubacterial and arc-haebaoterial kingdoms and these enzymes form a major link between protein and carbohydrate metabolism.
Reference Protein and Structure
- Sequence
-
P24295
 (1.4.1.2)
(1.4.1.2)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
[Clostridium] symbiosum (Bacteria)

- PDB
-
1hrd
- GLUTAMATE DEHYDROGENASE
(1.96 Å)



- Catalytic CATH Domains
-
3.40.50.10860
 (see all for 1hrd)
(see all for 1hrd)
Enzyme Reaction (EC:1.4.1.2)
Enzyme Mechanism
Introduction
The first step in the mechanism is the deprotonation of the alpha-amino group of the glutamate by Asp165, acting as general base, which occurs with subsequent hydride transfer to the Si face of the NAD+ and leads to the production of an iminoglutarate intermediate. The next step is the attack of a water molecule on the iminoglutarate intermediate is enhanced byLys125 acting as a general base via a hydrogen bond. During the generation of the carbinolamine intermediate and its subsequent collapse to the 2-oxoacid, Asp165 is crucial for the transfer of the proton to and from the substrate. Finally the loss of a proton from Lys125 and Asp165 ends the catalytic cycle.
Catalytic Residues Roles
| UniProt | PDB* (1hrd) | ||
| Lys126 | Lys125A | Low pKa of residue enable it to act as a hydrogen-bond acceptor to enhance the nucleophilicity of a water moelcule to enable attack on the iminoglutarate intermediate. | proton acceptor, electrostatic stabiliser, proton donor |
| Asp166 | Asp165A | General base in the initial deprotonation of the alpha-amino group of the glutamate. Acid/base catalyst during generation of 2-oxoacid intermediate and collapse. | proton acceptor, proton donor |
Chemical Components
proton transfer, aromatic bimolecular nucleophilic addition, overall reactant used, cofactor used, hydride transfer, bimolecular nucleophilic addition, intermediate formation, overall product formed, elimination (not covered by the Ingold mechanisms), deamination, inferred reaction stepReferences
- Dean JL et al. (1994), Biochem J, 301, 13-16. The catalytic role of aspartate in the active site of glutamate dehydrogenase. DOI:10.1042/bj3010013. PMID:8037659.
- Stillman TJ et al. (1993), J Mol Biol, 234, 1131-1139. Conformational Flexibility in Glutamate Dehydrogenase. DOI:10.1006/jmbi.1993.1665. PMID:8263917.
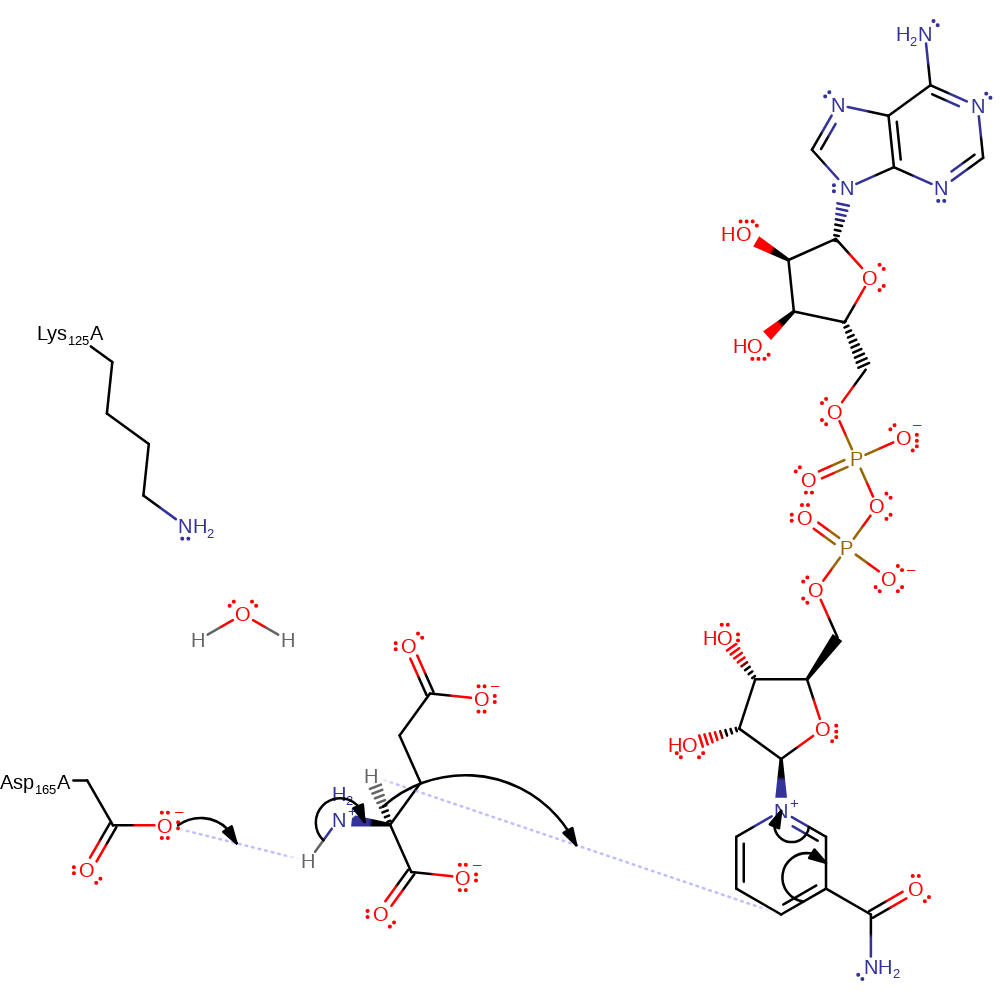
Step 1. Asp165 acts as a general base to deprotonate the amine group of the substrate. This occurs with subsequent hydride transfer to the Si face of NAD+ and leads to the formation of an iminoglutamate intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys125A | electrostatic stabiliser |
| Asp165A | proton acceptor |
Chemical Components
proton transfer, ingold: aromatic bimolecular nucleophilic addition, overall reactant used, cofactor used, hydride transfer
Step 2. Lys125 activates a water molecule for nucleophilic attack on the imine intermediate. Asp165 transfers a proton to the developing amine.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys125A | proton acceptor |
| Asp165A | proton donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation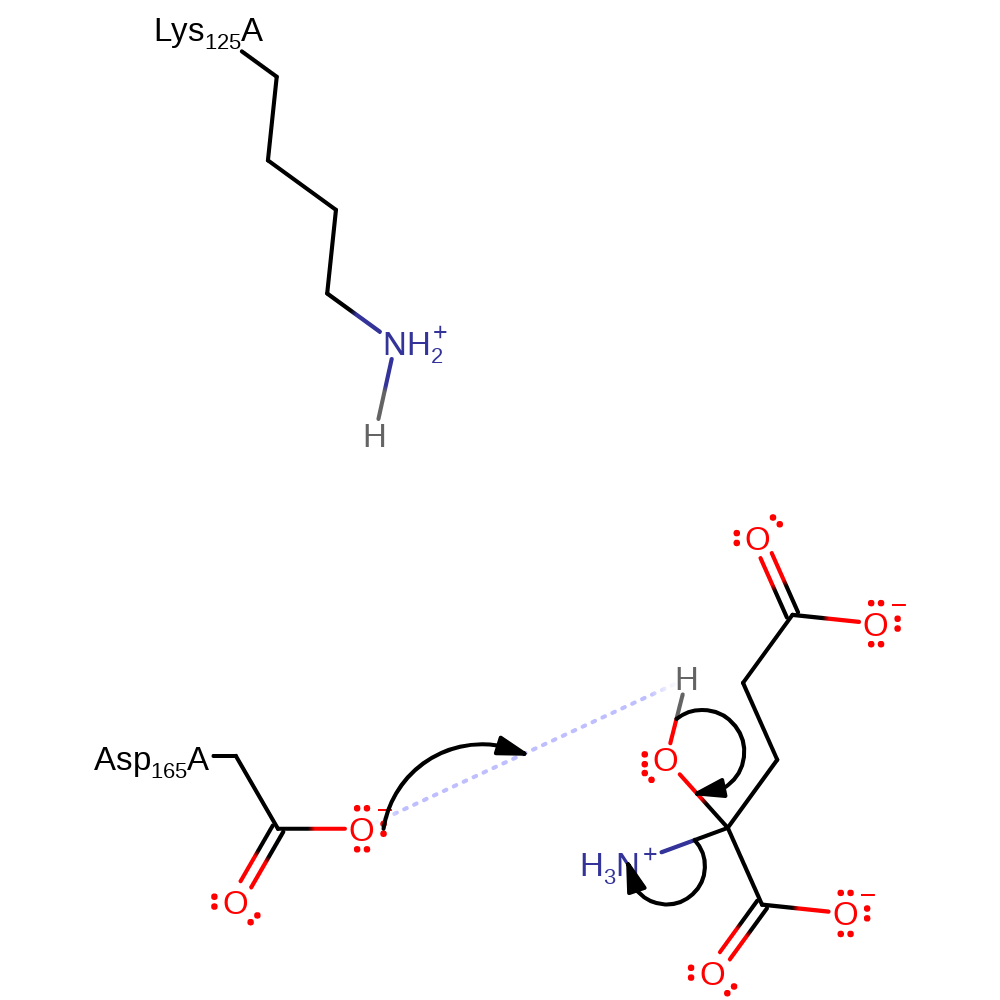
Step 3. Asp165 abstracts a proton from the hydroxyl group of the carbinolamine intermediate. The intermediate collapses and ammonia is eliminated.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp165A | proton acceptor |
Chemical Components
proton transfer, overall product formed, elimination (not covered by the Ingold mechanisms), deamination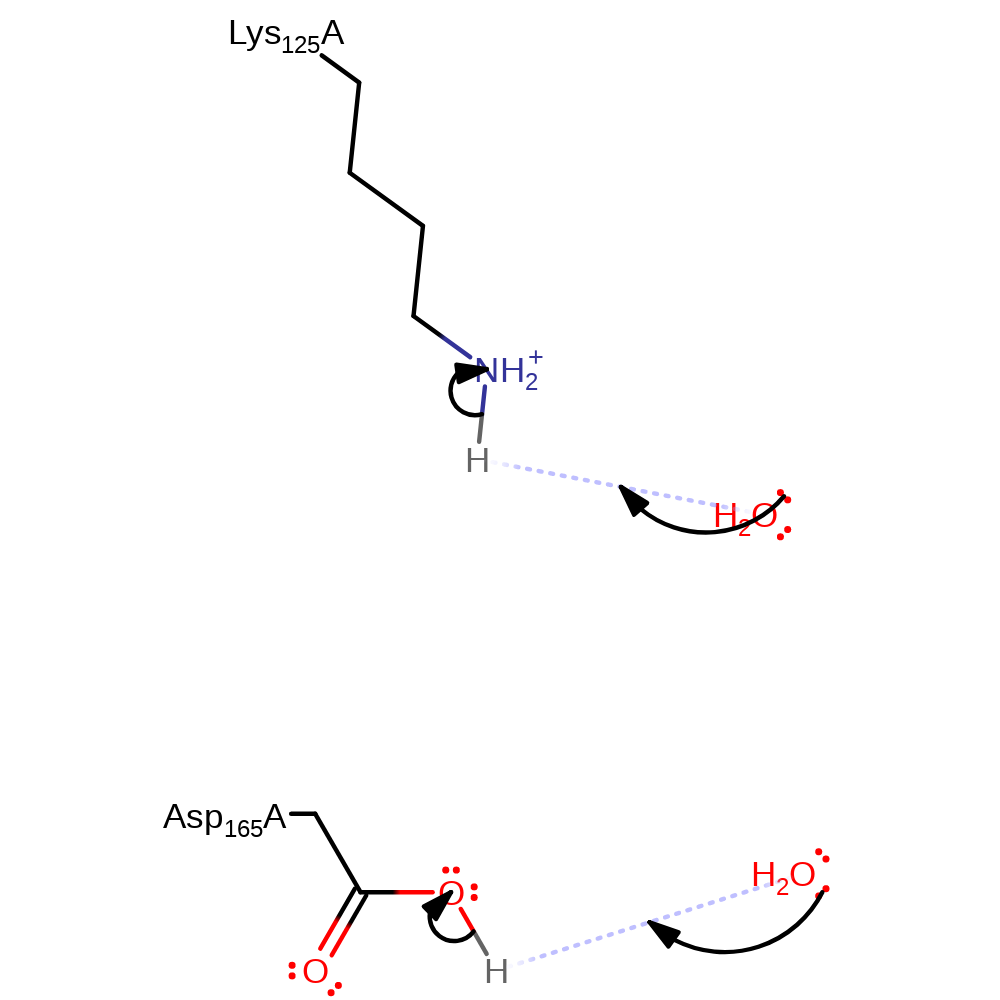
Step 4. In an inferred step Asp165 and Lys125 are deprotonated to regenerate the active site.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys125A | proton donor |
| Asp165A | proton donor |







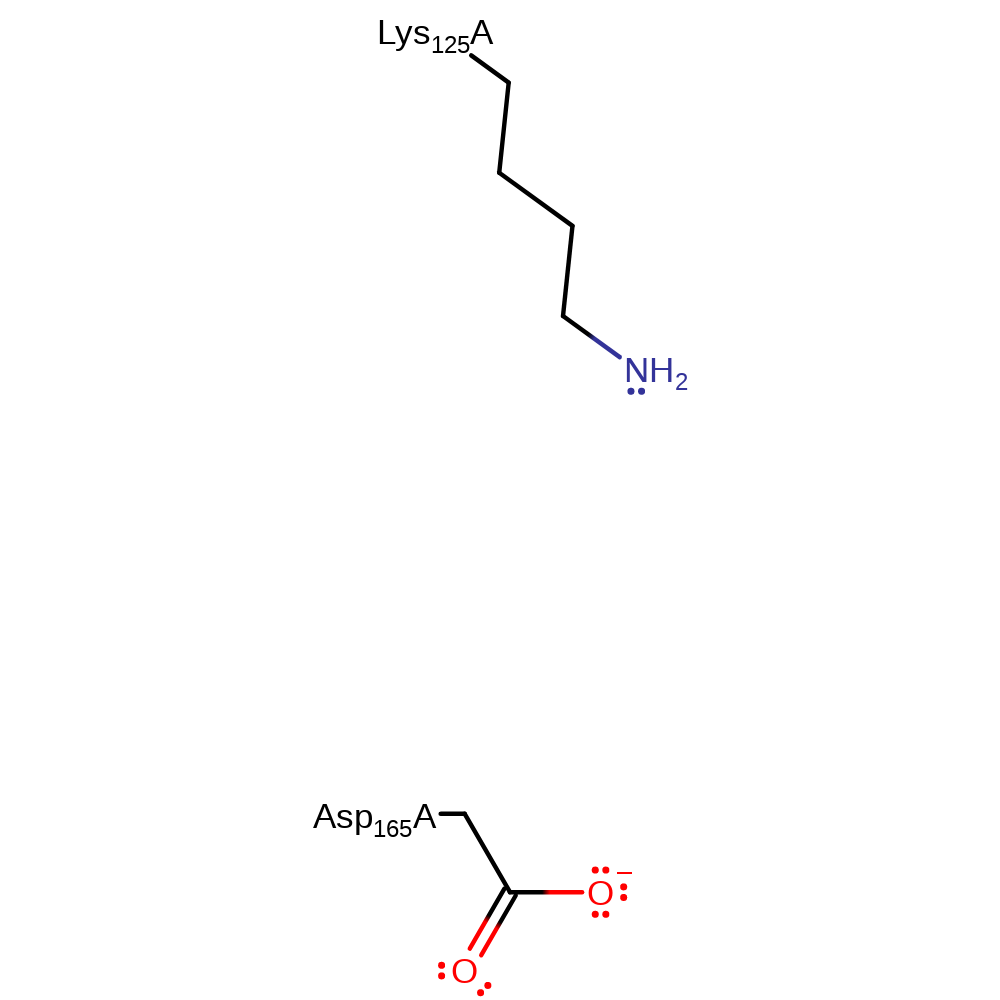 Download:
Download: