D-lactate dehydrogenase
D -Lactate dehydrogenase from Lactobacillus bulgaricus, a homodimer with 332 amino acid, acts at the last step of the glycolytic pathway under anaerobic conditions, allowing re-oxidation of NAD, which is necessary for glycolysis. The enzyme catalyses the NAD-dependent conversion of pyruvate into the D -isomer of lactic acid. The reaction is reversible: pyruvate reduction (the forward reaction) shows a maximum rate at pH 7.5 and D-lactate oxidation (the inverse reaction) at pH 8.0. The reaction leading to the other enantiomer, L-lactic acid, is catalysed by another enzyme, L-lactate dehydrogenase (L-LDH). While L-LDH has a wide occurrence in nature, D-LDH is found only in invertebrates, lower fungi and prokaryotic organisms. Lactic bacteria possess either one or both enzymes. In Lactobacillus bulgaricus, commonly used in the dairy industry for the production of yoghurt, more than 90% of the pyruvate is converted into D-lactate.
Reference Protein and Structure
- Sequence
-
P26297
 (1.1.1.28)
(1.1.1.28)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Lactobacillus delbrueckii subsp. bulgaricus ATCC 11842 = JCM 1002 (Bacteria)

- PDB
-
1j49
- INSIGHTS INTO DOMAIN CLOSURE, SUBSTRATE SPECIFICITY AND CATALYSIS OF D-LACTATE DEHYDROGENASE FROM LACTOBACILLUS BULGARICUS
(2.2 Å)



- Catalytic CATH Domains
-
3.40.50.720
 (see all for 1j49)
(see all for 1j49)
Enzyme Reaction (EC:1.1.1.28)
Enzyme Mechanism
Introduction
With reference to the known mechanism of L-lactate dehydrogenases, the reaction proceeds first with a proton being donated from (R)-lactate by His297 then a simultaneous proton donation to His297 and a hydride being transferred to NAD+, with help by Arg236 and Asp260 residues.
Catalytic Residues Roles
| UniProt | PDB* (1j49) | ||
| Asp260 | Asp260A | Asp260 twists the NAD+ carboxamide by 21 degrees from the pyridine plane and fixing its oxygen in the cis-orientation with respect to C4N. This conformation has been shown to be unfavourable in isolated NADH/NAD+ molecules, because of the partial double-bond character of the C3N±C4N bond, but it is important in the activation of hydride transfer in dehydrogenases. | steric role |
| Glu265 | Glu265A | H-bonds to His297 to stabilise the protonated form, increasing its pKa and aid in its function as a base in the concerted step | electrostatic stabiliser |
| His297 | His297A | Acts as a general acid/base and abstracts and donates a proton from pyruvate to form D-lactate. | proton acceptor, proton donor |
| Arg236 | Arg236A | Polarises the carbonyl bond in the pyruvate molecule to activate the hydride transfer. | electrostatic stabiliser |
Chemical Components
proton transfer, hydride transfer, aromatic bimolecular nucleophilic addition, overall reactant used, overall product formedReferences
- Razeto A et al. (2002), J Mol Biol, 318, 109-119. Domain Closure, Substrate Specificity and Catalysis of d-Lactate Dehydrogenase from Lactobacillus bulgaricus. DOI:10.1016/s0022-2836(02)00086-4. PMID:12054772.
- Kochhar S et al. (2000), Eur J Biochem, 267, 1633-1639. Roles of His205, His296, His303 and Asp259 in catalysis by NAD+-specific D-lactate dehydrogenase. DOI:10.1046/j.1432-1327.2000.01155.x. PMID:10712593.

Step 1. His297 abstracts a proton from the hydroxyl group of D-lactate. There is aromatic bimolecular nucleophilic addition of a hydride from the reactive carbon of D-lactate to NAD+, forming NADH and the product. Glu265 stabilises the protonated form of His297 and increases its pKa.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg236A | electrostatic stabiliser |
| Asp260A | steric role |
| Glu265A | electrostatic stabiliser |
| His297A | proton acceptor |
Chemical Components
proton transfer, hydride transfer, ingold: aromatic bimolecular nucleophilic addition, overall reactant used, overall product formed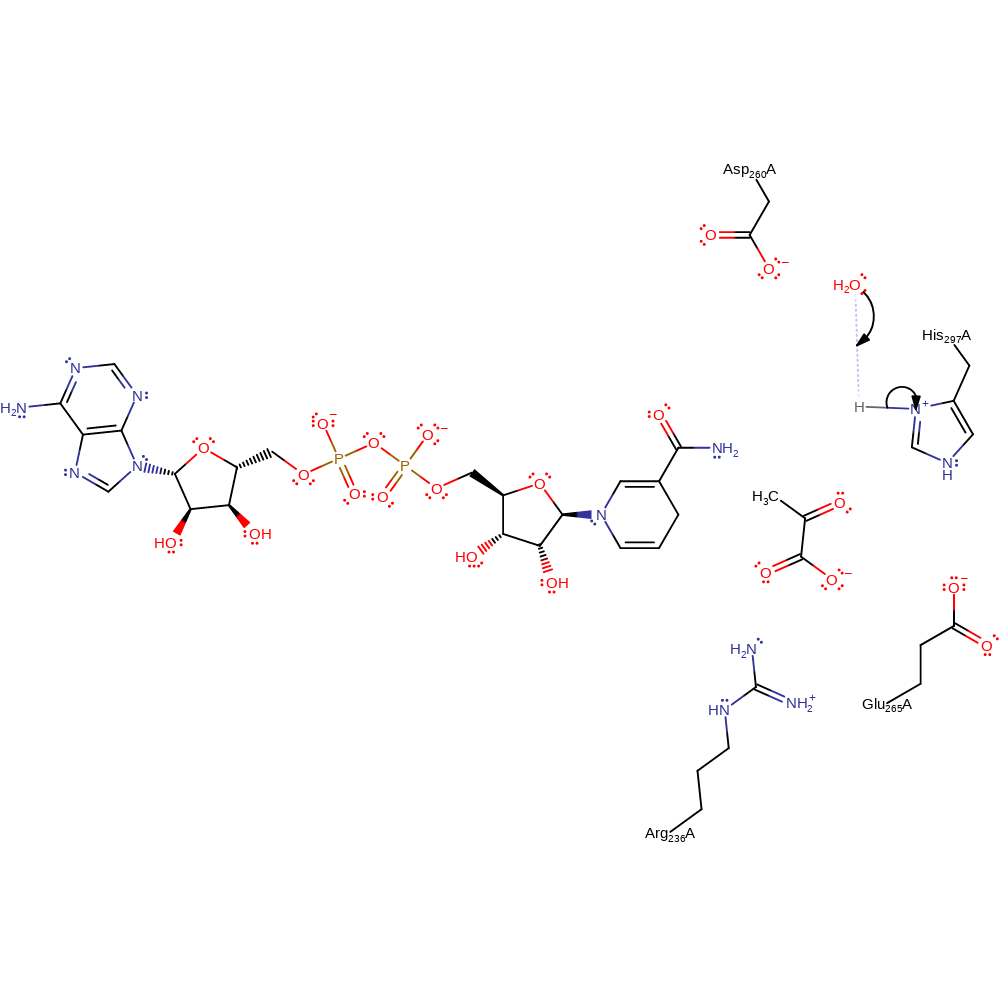
Step 2. In an inferred reaction step His297 is deprotonated to regenerate the native state of the enzyme.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His297A | proton donor |





 Download:
Download: