Lipopolysaccharide 3-alpha-galactosyltransferase
Lipo-polysaccharide galactosyl transferase (LGTC) is a retaining glycosyl transferase which catalyses the transfer of alpha-D-galactose to a terminal lactose during the synthesis of lipo-polysaccharides with retention of stereochemistry at the anomeric carbon. The enzyme held at the surface of the bacterial plasma membrane through electrostatic interaction between basic residues and phospho-lipids and interactions between hydrophobic regions.
Reference Protein and Structure
- Sequence
-
Q93EK7

 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Neisseria meningitidis (Bacteria)

- PDB
-
1ga8
- CRYSTAL STRUCTURE OF GALACOSYLTRANSFERASE LGTC IN COMPLEX WITH DONOR AND ACCEPTOR SUGAR ANALOGS.
(2.0 Å)



- Catalytic CATH Domains
-
3.90.550.10
 (see all for 1ga8)
(see all for 1ga8)
- Cofactors
- Magnesium(2+) (1)
Enzyme Reaction (EC:2.4.1.44)
Enzyme Mechanism
Introduction
It follows an SNi-type mechanism, similar to that of the OtsA enzyme. Lacking a strong nucleophile like other retaining enzymes in the glycosyltransferase 6 family (Gln189 is a poor nucleophile), O4 from lactose (galactose) directly attacks C1 on galactose in UDP-galactose front side via a dissociative SNi transition state. Proton transfer likely occurs from the 4'OH to O3 on the beta phosphate (UDP). The surrounding active site residues and Mn2+ ion help promote the front-side mechanism in the stabilisation of the reactant transition states. So far it is unclear whether a shortly lived intermediate like OtsA forms in the reaction and identify exactly the residues to stabilise the intermediate. Regardless, for LGTC it is far more favourable to proceed via a SNi mechanism, intermediate or not as opposed to the double displacement mechanism previously proposed.
Catalytic Residues Roles
| UniProt | PDB* (1ga8) | ||
| Lys250 | Lys250A | Lys250 out of all the residues has the largest electrostatic stabilisation effect by hydrogen bonds with the alpha and beta phosphates of the UDP leaving group. | hydrogen bond donor, electrostatic stabiliser, polar interaction |
| Asp103, Asp105, His244 | Asp103A, Asp105A, His244A | Co-ordinate to Mn2+ ion which in turn stabilises the negatively charged phosphate, making UDP a good leaving group. | metal ligand |
| Gln189 | Gln189A | Shown to have poor nucleophilicity, instead having a role in stabilising the developing charges during the transition state at the anomeric carbon in the reaction. | electrostatic stabiliser, polar interaction |
| His78, Asp130, Asp188 | His78A, Asp130A, Asp188A | Form hydrogen bonds with the reactants to stabilise the transition state. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic substitution, proton transfer, overall product formed, overall reactant usedReferences
- Gómez H et al. (2012), J Am Chem Soc, 134, 4743-4752. Retaining glycosyltransferase mechanism studied by QM/MM methods: lipopolysaccharyl-α-1,4-galactosyltransferase C transfers α-galactose via an oxocarbenium ion-like transition state. DOI:10.1021/ja210490f. PMID:22352786.
- Persson K et al. (2001), Nat Struct Biol, 8, 166-175. Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs. DOI:10.1038/84168. PMID:11175908.

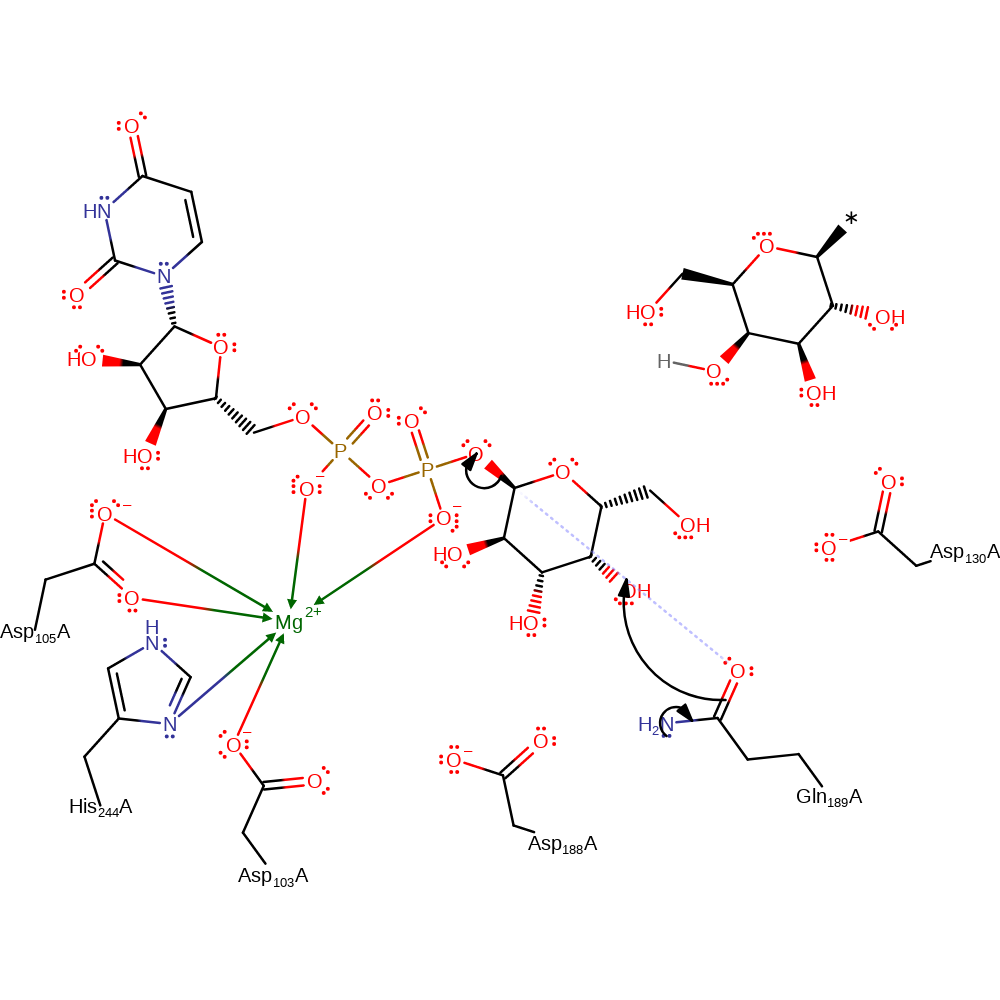
Step 1. Nucleophilic attack by 4'OH on glucose on the 1' anomeric C is facilitated by the protonation of the leaving group UDP.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp105A | metal ligand |
| His244A | metal ligand |
| Asp103A | metal ligand |
| Gln189A | electrostatic stabiliser |
| His78A | hydrogen bond donor |
| Asp130A | hydrogen bond acceptor |
| Asp188A | hydrogen bond acceptor |
| His78A | electrostatic stabiliser |
| Asp130A | electrostatic stabiliser |
| Asp188A | electrostatic stabiliser |
| Lys250A | electrostatic stabiliser, hydrogen bond donor, polar interaction |
| Gln189A | polar interaction |
Chemical Components
ingold: bimolecular nucleophilic substitution, proton transfer, overall product formed, overall reactant usedIntroduction
Structural data and comparisons to similar enzymes suggests a double displacement reaction, while computer modelling favours front side SNi attack. The first mechanism requires Gln189 to act as a nucleophile at the anomeric carbon, forming an enzyme-substrate imidic ester intermediate which is then attacked by the 4-OH group of the acceptor sugar. Inferred proton transfer to the UDP leaving group phosphate. Gln189 has subsequently been shown to be a weak nucleophile where a SNi mechanism is far more favoured with other enzymes also being shown to follow the SNi path.
Catalytic Residues Roles
| UniProt | PDB* (1ga8) | ||
| Gln189 | Gln189A | Experimental evidence has suggested that Gln189 uses its side chain nitrogen as a nucleophile at the anomeric carbon of the alpha-D-galactose substrate. The enzyme-substrate imidic ester intermediate is then attacked by the 4-OH of the acceptor lactose, forming the product with retention of stereochemistry. However, no intermediate has been observed for this enzyme. | covalently attached, nucleofuge, nucleophile |
| Asp130, Asp188 | Asp130A, Asp188A | The residue's side chain stabilises the transition state. | electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic substitution, enzyme-substrate complex formation, intermediate formation, overall reactant used, proton transfer, enzyme-substrate complex cleavage, overall product formed, intermediate collapse, inferred reaction step, native state of enzyme regeneratedReferences
- Persson K et al. (2001), Nat Struct Biol, 8, 166-175. Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs. DOI:10.1038/84168. PMID:11175908.
- Tvaroska I (2004), Carbohydr Res, 339, 1007-1014. Molecular modeling insights into the catalytic mechanism of the retaining galactosyltransferase LgtC. DOI:10.1016/j.carres.2003.11.014. PMID:15010308.
Step 1. Gln189 carbonyl acts as a nucleophile and attacks C1 on UDP-galactose to form a covalent intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln189A | covalently attached |
| Asp188A | electrostatic stabiliser |
| Asp130A | electrostatic stabiliser |
| Asp103A | metal ligand |
| Asp105A | metal ligand |
| His244A | metal ligand |
| Gln189A | nucleophile |
Chemical Components
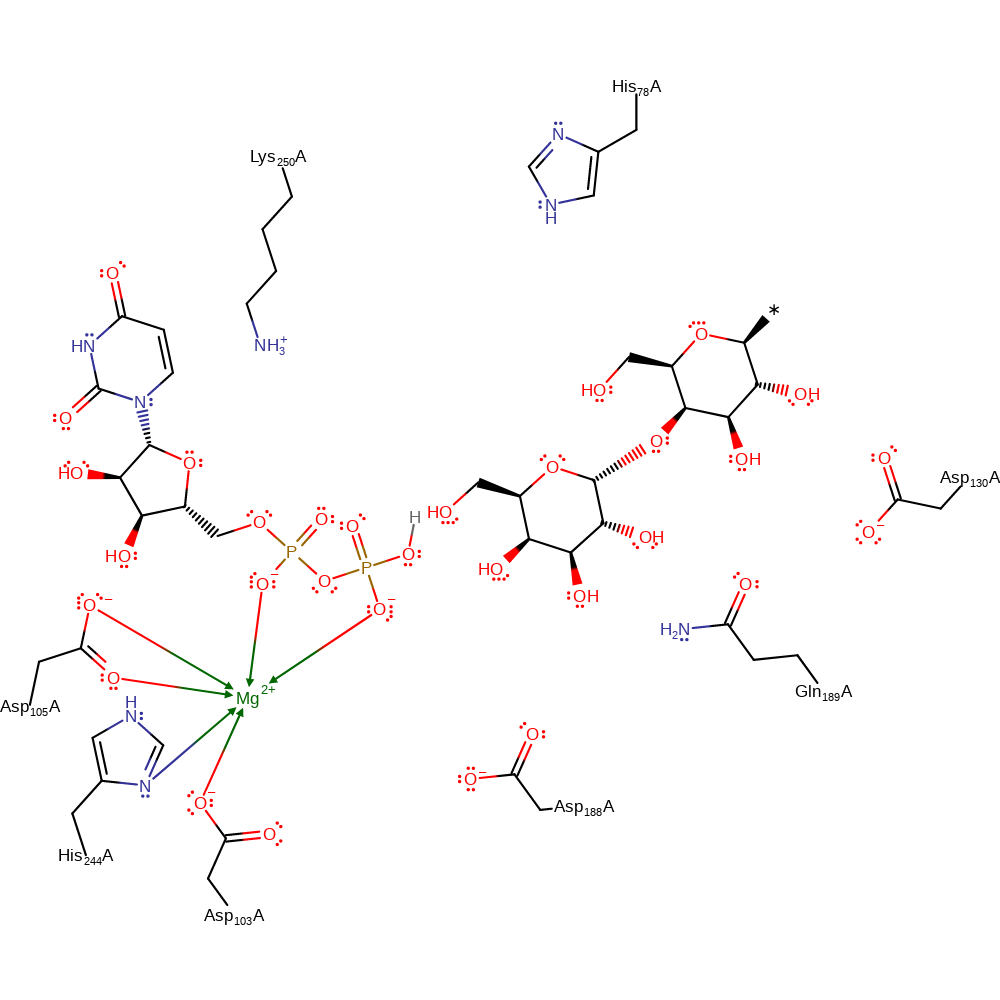
ingold: bimolecular nucleophilic substitution, enzyme-substrate complex formation, intermediate formation, overall reactant used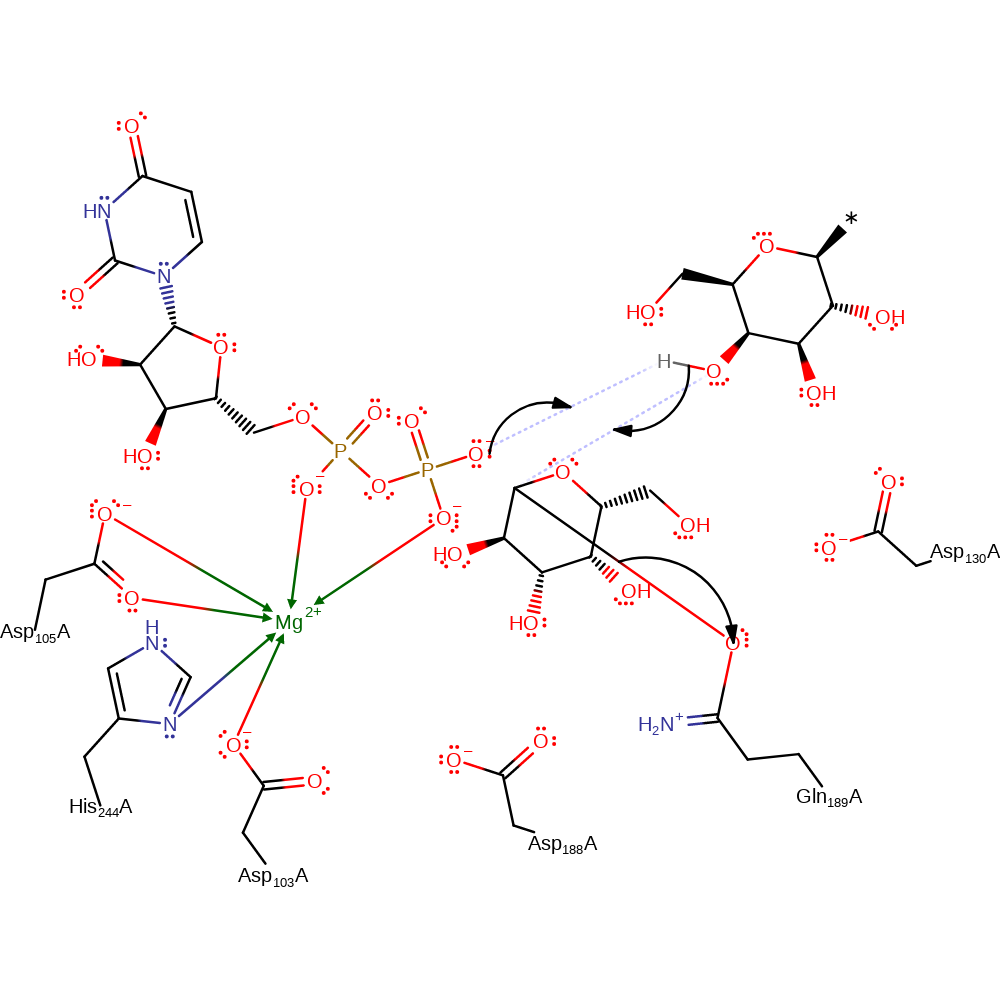
Step 2. Nucleophilic attack of the 4'OH on the enzyme-substrate intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln189A | covalently attached |
| Asp103A | metal ligand |
| Asp105A | metal ligand |
| His244A | metal ligand |
| Gln189A | nucleofuge |





 Download:
Download: 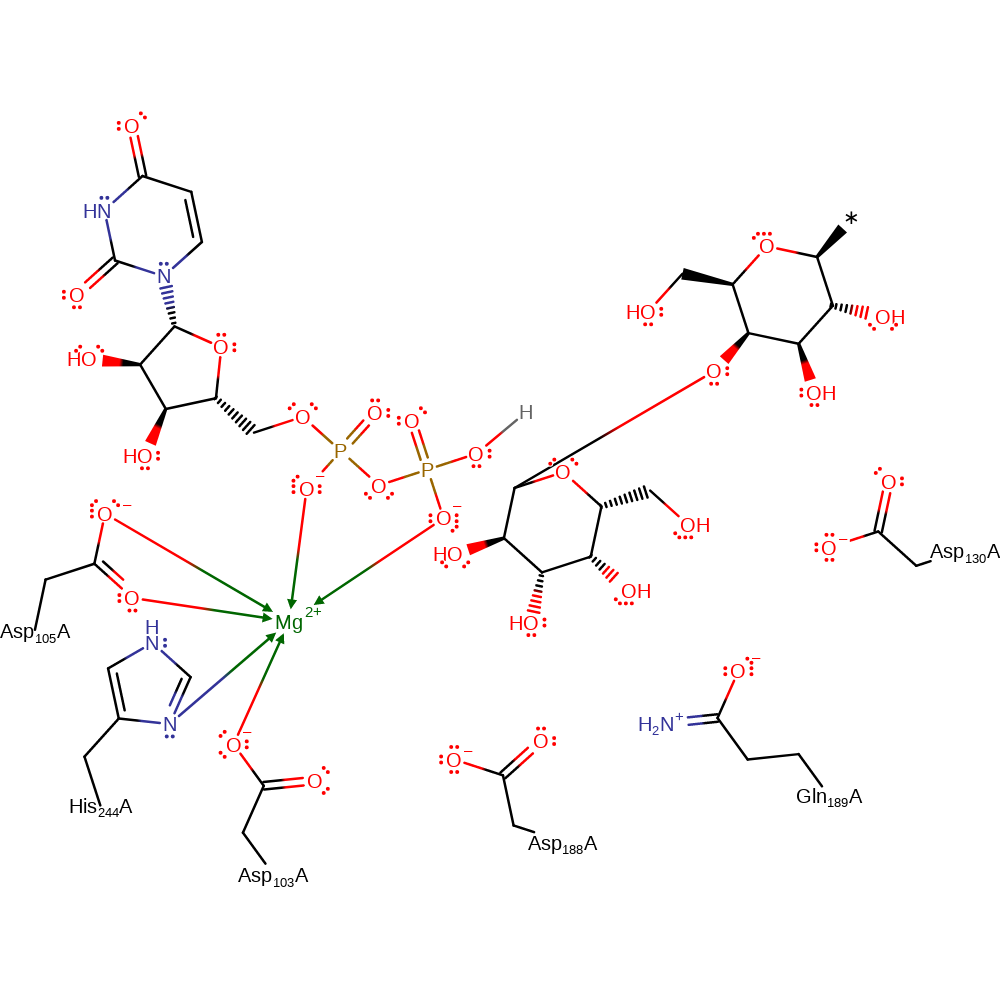 Download:
Download: