Glucose oxidase
Glucose oxidase is a flavin dependent glycoprotein. The fungal enzyme is a homodimer made up of two identical subunits, each containing one molecule of non-covalently bound FAD. The enzyme catalyses the oxidation of beta-D-glucose, where the FAD cofactor acts as the redox carrier.
Reference Protein and Structure
- Sequence
-
P13006
 (1.1.3.4)
(1.1.3.4)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Aspergillus niger (Fungus)

- PDB
-
1gal
- CRYSTAL STRUCTURE OF GLUCOSE OXIDASE FROM ASPERGILLUS NIGER: REFINED AT 2.3 ANGSTROMS RESOLUTION
(2.3 Å)



- Catalytic CATH Domains
-
3.30.560.10
 (see all for 1gal)
(see all for 1gal)
- Cofactors
- Fadh2(2-) (1)
Enzyme Reaction (EC:1.1.3.4)
Enzyme Mechanism
Introduction
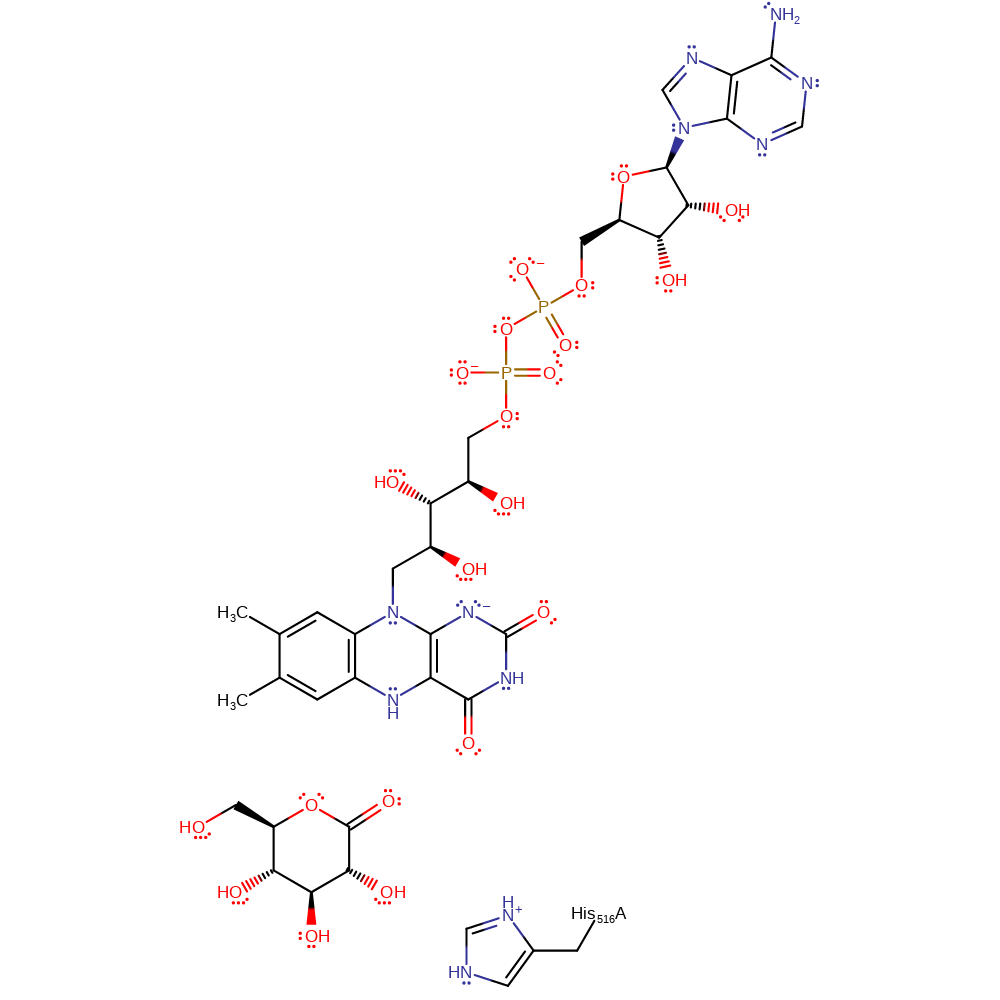
Crystallographic investigations have shown the flavin containing active site to be buried in a deep pocket. The binding of glucose to the free enzyme results in the expulsion of a water molecule and a proton from the active site.
In the reduction half reaction, simultaneous hydride and proton transfer from the glucose to the FAD and His516 respectively occurs.
In the oxidation half reaction, the reduced coenzyme FADH- is reoxidised back to FAD by molecular dioxygen which undergoes reduction to hydrogen peroxide in two single electron transfer steps.The hydride transfer is thought to be regulated by dissociation between Glu412 and His559. The electrostatic interaction between these residues controls the pH and therefore reactivity of the active site, although they are not directly involved in the catalytic mechanism.
Catalytic Residues Roles
| UniProt | PDB* (1gal) | ||
| His538 | His516A | The residue acts as a general base towards the C1-OH group of the glucose substrate. As the proton is removed, the oxygen partial negative charge drives the hydride transfer to the N5 of FAD resulting in FADH-. A ketone is formed at the C1 of glucose. | proton acceptor |
Chemical Components
hydride transfer, proton transfer, native state of cofactor regenerated, overall reactant used, aromatic bimolecular nucleophilic addition, overall product formedReferences
- Leskovac V et al. (2005), Int J Biochem Cell Biol, 37, 731-750. Glucose oxidase from Aspergillus niger: the mechanism of action with molecular oxygen, quinones, and one-electron acceptors. DOI:10.1016/j.biocel.2004.10.014. PMID:15694834.
- Wang Y et al. (2018), Chem Sci, 9, 594-599. FAD roles in glucose catalytic oxidation studied by multiphase flow of extractive electrospray ionization (MF-EESI) mass spectrometry. DOI:10.1039/C7SC04259K. PMID:29629123.
- Hecht HJ et al. (1993), J Mol Biol, 229, 153-172. Crystal structure of glucose oxidase from Aspergillus niger refined at 2.3 A resolution. PMID:8421298.

Step 1. In the reductive half-reaction there is concerted transfer of a proton from glucose to His516 and a hydride from glucose to FAD, forming FADH- and the product. FADH- is reoxidized by oxygen in the oxidative half-reaction. This proceeds via two single-electron transfer steps and involves superoxide anion radical and flavin semiquinone radical intermediates.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His516A | proton acceptor |




 Download:
Download: