Formate dehydrogenase
Formate Dehydrogenase H (FDH-H) from Escherichi coli is a 79kDa polypeptide component of the anaerobic formate hydrogen lyase complex. The enzyme catalyses the oxidation of formate (produced from pyruvate during anaerobic growth) to carbon dioxide, in the absence of exogenous electron acceptors, with the concomitant release of two electrons and two protons.
Reference Protein and Structure
- Sequence
-
P07658
 (1.17.98.4)
(1.17.98.4)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1aa6
- REDUCED FORM OF FORMATE DEHYDROGENASE H FROM E. COLI
(2.3 Å)



- Catalytic CATH Domains
-
3.40.228.10
 2.20.25.90
2.20.25.90  (see all for 1aa6)
(see all for 1aa6)
- Cofactors
- Molybdenum(6+) (1), Tetra-mu3-sulfido-tetrairon (1), Molybdopterin guanine dinucleotide (2)
Enzyme Reaction (EC:1.17.1.9)
Enzyme Mechanism
Introduction
Initially, formate binds to Mo 800 via one of its oxygens and displaces SeCys140. Formate is then oxidised to carbon dioxide. Two electrons are transferred to Mo800 reducing it from Mo(VI) to Mo(IV). The transfer of electrons from formate to Mo 800 may occur by direct two-electron transfer or by direct hydride transfer. The alpha proton of formate is transferred to SeCys140 and then to His141. A selenium-carboxylated intermediate may be formed before CO2 is released. Electrons from Mo(IV) are shuttled to the Fe4S4 cluster one at a time so an intermediate with Mo(V) and reduced Fe4S4 is produced. This is supported by evidence from electron paramagnetic resonance (EPR) experiments. Electron shuttling occurs by a ping pong mechanism through the partly delocalised ring of molybdopterin guanine dinucleotide (MGD) 802 and hydrogen bonds linking MGD, HOH 30, Lys44 and the Fe4S4 cluster. The Fe4S4 cluster is then oxidised by another electron acceptor. It is not known what functions as the final electron acceptor in vivo. Benzyl viologen (BV) has been used as the final electron acceptor in experiments. After Mo(V) is oxidised to Mo(IV) the proton on His141 can be released to the solvent.
Catalytic Residues Roles
| UniProt | PDB* (1aa6) | ||
| Arg333 | Arg333A | Stabilises the free selenol on SeCys140 after it is displaced from Mo by the substrate. | electrostatic stabiliser |
| His141 | His141A | After the proton from formate is transferred to SeCys140 it is passed on to His141. Evidence from electron paramagnetic resonance also supports the protonation of His141 by the alpha proton of formate (PMID: 9036855). | proton acceptor, electrostatic stabiliser |
| Lys44 | Lys44A | Facilitates electron transfer between HOH 30 and the Fe4S4 cluster. | electrostatic stabiliser |
| Sec140 | Sec140A | Accepts the alpha proton from formate. A selenium-carboxylated intermediate may be formed on this residue during the oxidation of formate to CO2. | nucleofuge, nucleophile, metal ligand, proton acceptor, proton donor |
Chemical Components
coordination to a metal ion, overall product formed, overall reactant used, electron transfer, redox reaction, electron relay, native state of enzyme regeneratedReferences
- Boyington JC et al. (1997), Science, 275, 1305-1308. Crystal Structure of Formate Dehydrogenase H: Catalysis Involving Mo, Molybdopterin, Selenocysteine, and an Fe4S4 Cluster. DOI:10.1126/science.275.5304.1305. PMID:9036855.
- Leopoldini M et al. (2008), Chemistry, 14, 8674-8681. Reaction mechanism of molybdoenzyme formate dehydrogenase. DOI:10.1002/chem.200800906. PMID:18671310.
- Raaijmakers HC et al. (2006), J Biol Inorg Chem, 11, 849-854. Formate-reduced E. coli formate dehydrogenase H: the reinterpretation of the crystal structure suggests a new reaction mechanism. DOI:10.1007/s00775-006-0129-2. PMID:16830149.
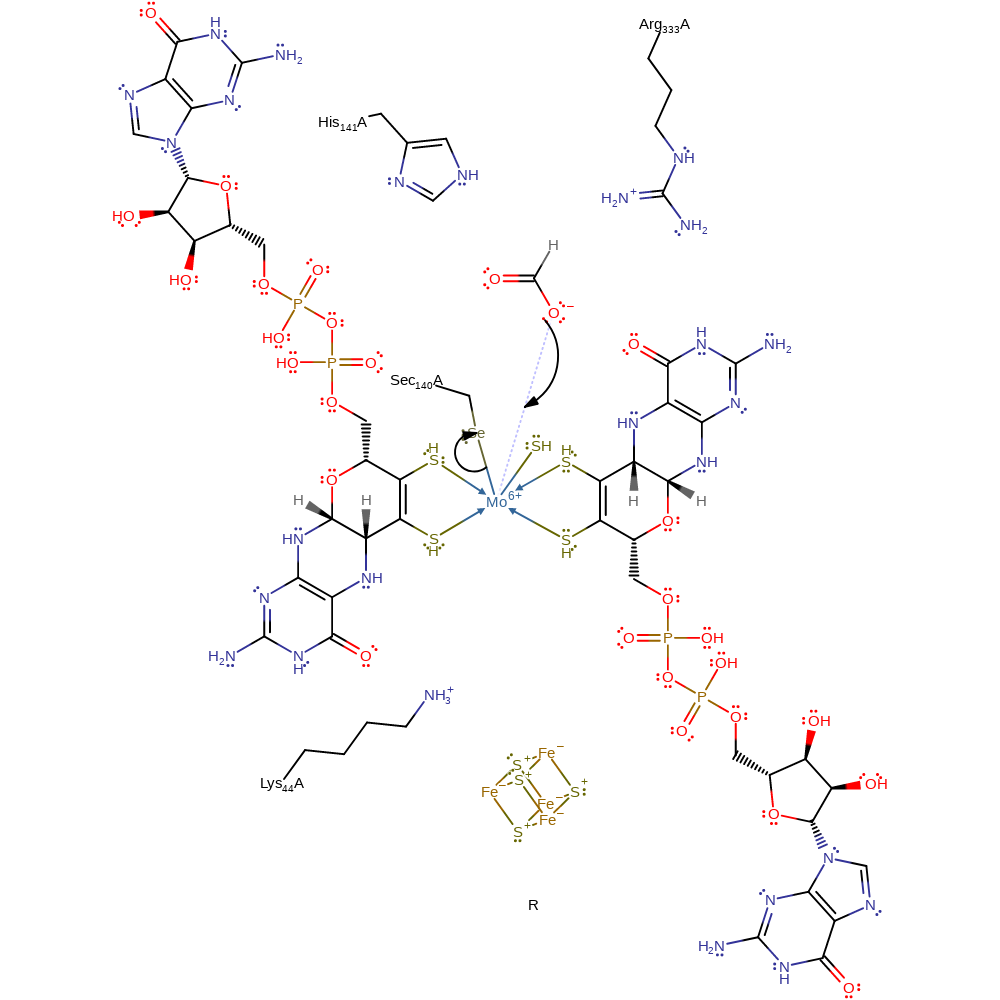
Step 1. Catalysis begins with the formate displacing the Mo-bound Sec140 group in the oxidised form of the enzyme.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg333A | electrostatic stabiliser |
| Lys44A | electrostatic stabiliser |
| His141A | electrostatic stabiliser |
| Sec140A | metal ligand |
| Sec140A | nucleofuge |
Chemical Components
coordination to a metal ion
Step 2. The formate proton is abstracted by Sec140. The CO2 molecule is released and two electrons are transferred to Mo, forming Mo(IV).
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His141A | electrostatic stabiliser |
| Arg333A | electrostatic stabiliser |
| Sec140A | proton acceptor |
Chemical Components
overall product formed, overall reactant used, electron transfer, redox reaction
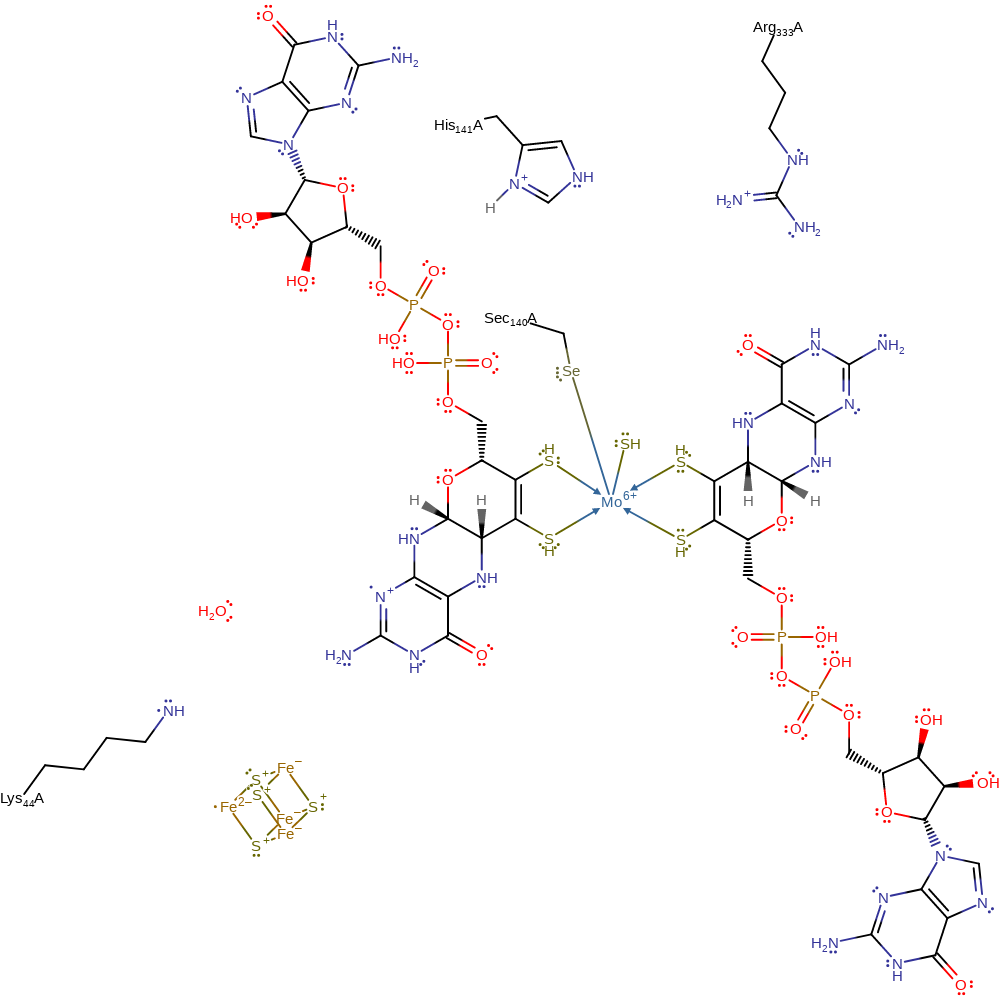
Step 3. An electron is shuttled from Mo(IV) to an electron acceptor (R) through an MGD ligand, a water molecule, then the Fe4S4 cluster. This results in a Mo(V) intermediate. The formate proton is transferred from Sec140 to His141. This leads to hydrogen bond formation between His141 and Sec140.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His141A | electrostatic stabiliser, proton acceptor |
| Sec140A | proton donor |
Chemical Components
electron transfer, electron relay, electron transfer, redox reaction
Step 4. Once the Fe4S4 cluster is reoxidized, a second electron is transferred from Mo(V) to the Fe4S4 cluster. The oxidation of Mo(V) to Mo(VI) causes the hydrogen bond between Sec140 and His141 to break, releasing the proton of His141 to solvent. Sec140 re-cordinates to the Mo(VI) centre. After the Fe4S4 cluster is reoxidised for a second time, the enzyme is restored to its initial state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Sec140A | nucleophile |
Chemical Components
native state of enzyme regenerated, electron relay, redox reactionIntroduction
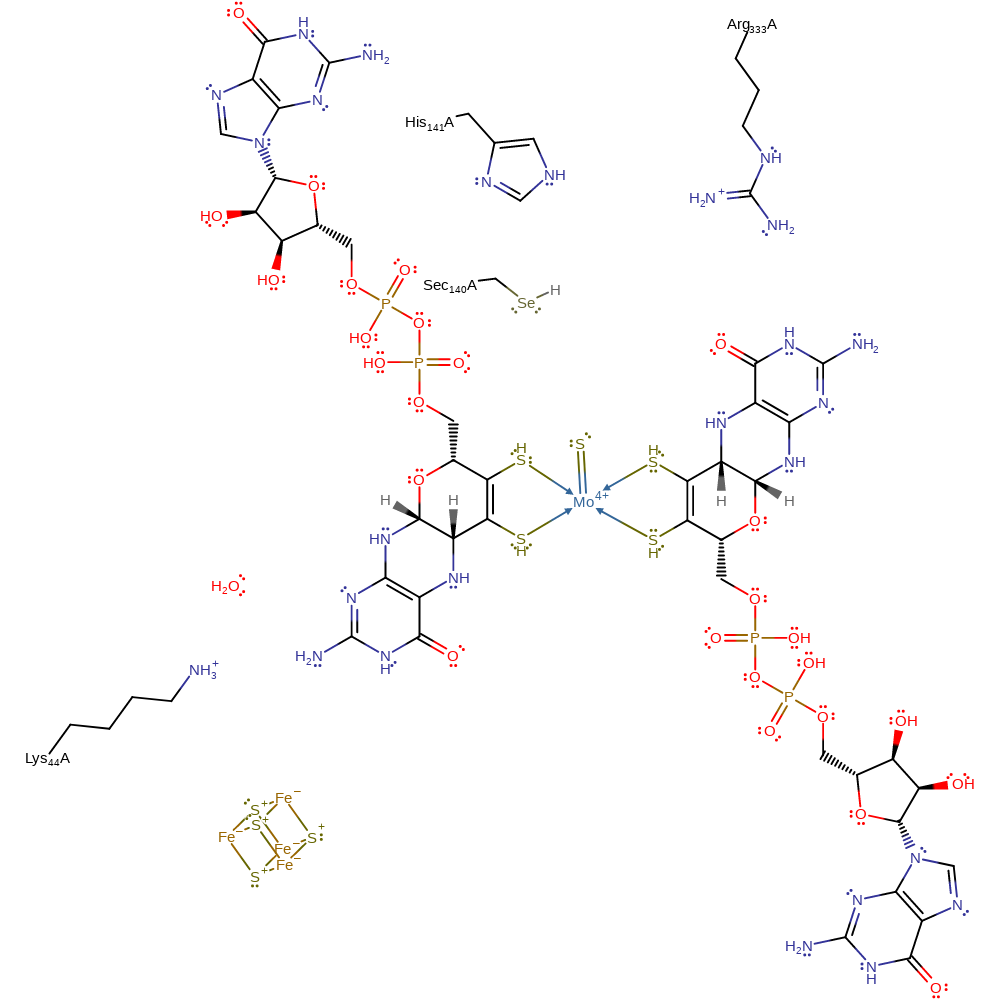
The hydrogen of the SH ligand on the Mo(VI) centre is abstracted by the Sec140 residue. There is direct hydride transfer from formate to Mo(VI), releasing CO2 and forming a Mo-H intermediate. The proton is then removed by the sulphur anion, reducing Mo(VI) to Mo(IV). The native state of the enzyme is restored when two electrons are transferred to an electron acceptor through the MGD ligands and Fe4S4 cluster, and Sec140 is deprotonated.
Catalytic Residues Roles
| UniProt | PDB* (1aa6) | ||
| Arg333 | Arg333A | Arg333 interacts with the Sec140 ligand and forms a hydrogen bond to formate. | hydrogen bond donor, electrostatic stabiliser |
| Sec140 | Sec140A | In this proposal Sec140 is not initially coordinated to the Mo(VI) centre. Sec140 deprotonates the sulphur ligand attached to Mo. | proton acceptor |
Chemical Components
proton transfer, hydride transfer, overall reactant used, overall product formed, bimolecular elimination, redox reaction, electron transfer, native state of enzyme regenerated, electron relayReferences
- Tiberti M et al. (2012), Inorg Chem, 51, 8331-8339. Evidence for the Formation of a Mo–H Intermediate in the Catalytic Cycle of Formate Dehydrogenase. DOI:/10.1021/ic300863d.
- Hartmann T et al. (2015), Biochim Biophys Acta, 1854, 1090-1100. Assembly and catalysis of molybdenum or tungsten-containing formate dehydrogenases from bacteria. DOI:10.1016/j.bbapap.2014.12.006. PMID:25514355.

Step 1. The selenide ion of Sec140 abstracts a proton from the Mo-bound sulphur. Arg333 stabilises unprotonated Sec140 and forms a hydrogen bond with the oxygen of coordinated formate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg333A | electrostatic stabiliser, hydrogen bond donor |
| Sec140A | proton acceptor |
Chemical Components
proton transfer
Step 2. B-elimination of a hydride from formate to the Mo(VI) centre occurs and CO2 is released.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
hydride transfer, overall reactant used, overall product formed, ingold: bimolecular elimination
Step 3. Proton transfer to the Mo-bound sulphur ion occurs, reducing the Mo centre to Mo(IV).
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
proton transfer, redox reaction, electron transfer
Step 4. The active site is restored when two electrons are transferred from Mo(IV) to the electron acceptor (R) and Sec140 is deprotonated.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
redox reaction, native state of enzyme regenerated, electron relayIntroduction
A computational study of the mechanism of formate dehydrogenase proposed that the inactive state of the enzyme contains a partial bond between the Mo-bound sulphur ligand and Sec140. An activation step occurs before catalysis involving a 'sulphur shift'. Firstly, formate binds to the sulphur ligand and is stabilised by the positive charge of Arg333. The 'sulphur shift' then occurs as formate coordinates to the Mo centre and displaces Sec140 which binds to sulphur with a single bond. The sulphur-selenium bond breaks and the resulting selenide ion abstracts the proton from formate. His141 stabilises the free selenide with hydrogen bonds and lowers the activation energy for proton abstraction. CO2 then forms an additional bond to the sulphur. Product release occurs when the Mo-oxygen bond breaks, inducing the cleavage of the sulphur-carbon bond. Two electrons are transferred from the reduced Mo(IV) centre to an unknown electron acceptor, via Lys44 and the Fe4S4 cluster. The active site is regenerated when Sec140 is deprotonated and recoordinates to Mo(VI). However, this proposal does not include a Mo(V) intermediate during formate oxidation which has been identified by EPR studies.
Catalytic Residues Roles
| UniProt | PDB* (1aa6) | ||
| Arg333 | Arg333A | The positive charge of Arg333 stabilises the formate anion in the initial binding at the Mo(VI) centre. | electrostatic stabiliser |
| His141 | His141A | Forms a hydrogen bond with the Sec140 anion which lowers the activation energy for proton abstraction from formate. | |
| Lys44 | Lys44A | Involved in electron shuttling during the re-oxidation of Mo(IV) to Mo(VI). | |
| Sec140 | Sec140A | Sec140 is displaced from the active site following formate coordination and the 'sulphur shift'. The selenide ion deprotonates formate. | nucleofuge, metal ligand, proton acceptor, proton donor, electrophile |
Chemical Components
decoordination from a metal ion, coordination to a metal ion, proton transfer, overall product formed, native state of enzyme regenerated, redox reactionReferences
- Mota CS et al. (2011), J Biol Inorg Chem, 16, 1255-1268. The mechanism of formate oxidation by metal-dependent formate dehydrogenases. DOI:10.1007/s00775-011-0813-8. PMID:21773834.
- Hartmann T et al. (2015), Biochim Biophys Acta, 1854, 1090-1100. Assembly and catalysis of molybdenum or tungsten-containing formate dehydrogenases from bacteria. DOI:10.1016/j.bbapap.2014.12.006. PMID:25514355.
- Boyington JC et al. (1997), Science, 275, 1305-1308. Crystal Structure of Formate Dehydrogenase H: Catalysis Involving Mo, Molybdopterin, Selenocysteine, and an Fe4S4 Cluster. DOI:10.1126/science.275.5304.1305. PMID:9036855.

Step 1. In the inactive form of the enzyme there is a partial bond between the Mo sulphur ligand and the Sec140 residue. Catalysis beings when the oxygen atom of formate binds to the sulphur. The formate is stabilised by Arg333.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Sec140A | metal ligand |
| Arg333A | electrostatic stabiliser |
Chemical Components

Step 2. A sulphur shift occurs, in which the sulphur replaces the position of the Sec140 ligand, causing it to dissociate and form a covalently bound Sec140 intermediate to sulhpur. Formate binds to Mo(VI) via a single bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Sec140A | electrophile, nucleofuge |
Chemical Components
decoordination from a metal ion, coordination to a metal ionCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Sec140A | nucleofuge |
Chemical Components

Step 4. His141 stabilises the free selenide. The selenide ion abstracts the formate proton and the remaining CO2 molecule forms and additional bond to the sulphur ligand.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Sec140A | proton acceptor |
Chemical Components
proton transfer, overall product formed
Step 5. Product release occurs prior to the reoxidation of Mo(IV). The Mo-oxygen bond breaks, which induces the break of the sulphur-carbon bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
overall product formed
Step 6. Two electrons are transferred from Mo(IV) to an electron acceptor (R) via Lys44 and the Fe4S4 cluster. The proton on Sec140 is likely to be removed by solvent. After deprotonation, the Sec140 residue rebinds to the Mo(VI) centre and the active site is regenerated.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Sec140A | metal ligand, proton donor |





 Download:
Download:  Download:
Download: 
 Download:
Download: