Cellulase (GH12)
The glycosyl hydrolase family 12 member CelB is able to catalyse the breakdown of cellulose into glucose with retention of anomeric configuration. As a result, it is part of the endoglucanase clan GH-C which also includes the family 11 Xylanases, with a common fold and catalytic mechanism defining the clan.
Reference Protein and Structure
- Sequence
-
Q54331

 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Streptomyces lividans (Bacteria)

- PDB
-
2nlr
- STREPTOMYCES LIVIDANS ENDOGLUCANASE (EC: 3.2.1.4) COMPLEX WITH MODIFIED GLUCOSE TRIMER
(1.2 Å)



- Catalytic CATH Domains
-
2.60.120.180
 (see all for 2nlr)
(see all for 2nlr)
Enzyme Reaction (EC:3.2.1.4)
Enzyme Mechanism
Introduction
The reaction proceeds via a double displacement mechanism. Initial attack of the nucleophilic residue Glu 120 on the anomeric carbon of the beta-1-4-glycosidic bond results in cleavage of the bond, assisted by protonation of the leaving group at the 1C OH by Glu 203. This leads to a covalently bound enzyme intermediate, hydrolysis of which is achieved by a water molecule activated by deprotonation by Glu 203, releasing the products and regenerating the catalytically active forms of the nucleophilic and acid-base glutamates.
Catalytic Residues Roles
| UniProt | PDB* (2nlr) | ||
| Glu243 | Glu203A | Protonates the leaving group in the first stage and activates the water for hydrolysis in the second stage. | proton acceptor, proton donor, activator, increase nucleophilicity, promote heterolysis |
| Glu160 | Glu120A | Acts as nucleophile to attack the anomeric carbon resulting in the formation of a covalently bound enzyme intermediate. | covalently attached, nucleofuge, nucleophile |
Chemical Components
overall product formed, overall reactant used, bimolecular nucleophilic substitution, proton transfer, intermediate formation, hydrolysis, intermediate terminatedReferences
- Sulzenbacher G et al. (1999), Biochemistry, 38, 4826-4833. The Crystal Structure of a 2-Fluorocellotriosyl Complex of theStreptomyceslividansEndoglucanase CelB2 at 1.2 Å Resolution†,‡. DOI:10.1021/bi982648i. PMID:10200171.
- Crennell SJ et al. (2006), J Mol Biol, 356, 57-71. Dimerisation and an Increase in Active Site Aromatic Groups as Adaptations to High Temperatures: X-ray Solution Scattering and Substrate-bound Crystal Structures of Rhodothermus marinus Endoglucanase Cel12A. DOI:doi:10.1016/j.jmb.2005.11.004.
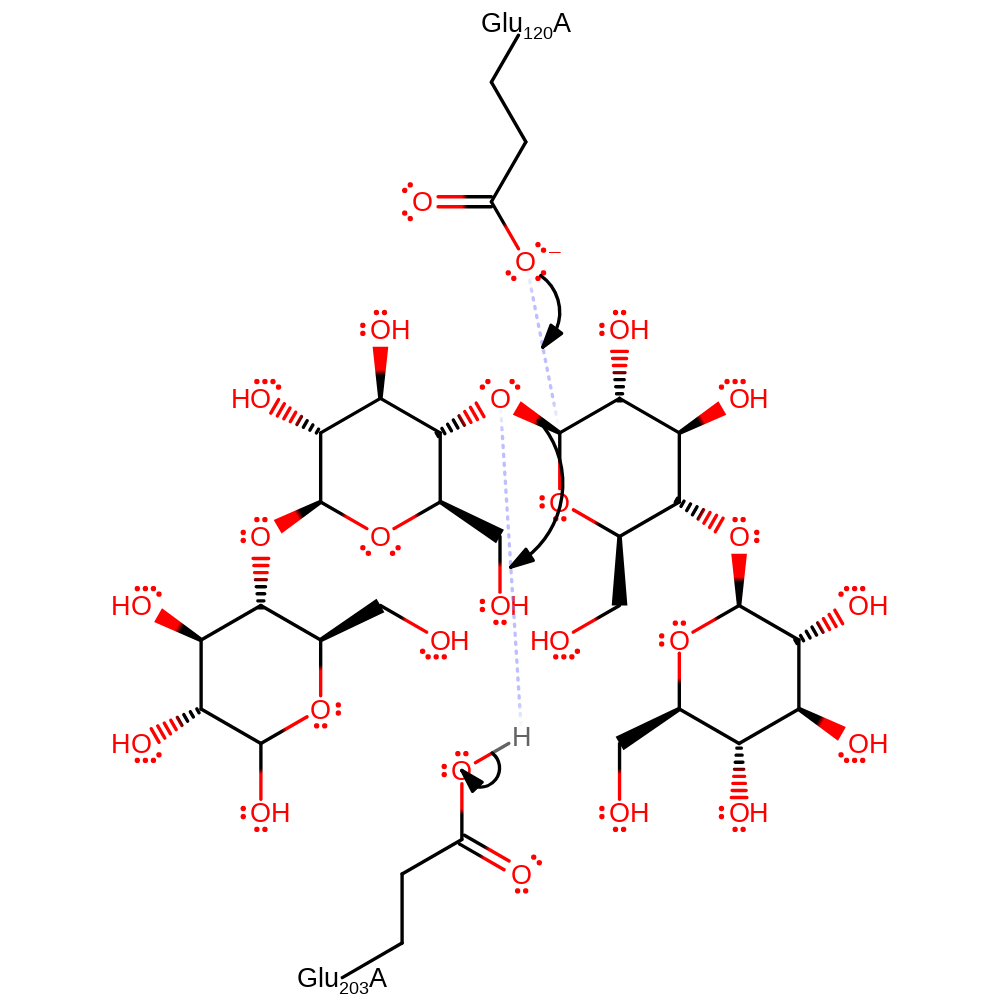
Step 1. Glu120 performs a nucleophilic attack on the anomeric carbon this causes the cleavage of the glycosidic bond assisted by Glu203 protonating the leaving group.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu120A | covalently attached |
| Glu203A | promote heterolysis, proton donor |
| Glu120A | nucleophile |
Chemical Components
overall product formed, overall reactant used, ingold: bimolecular nucleophilic substitution, proton transfer, intermediate formation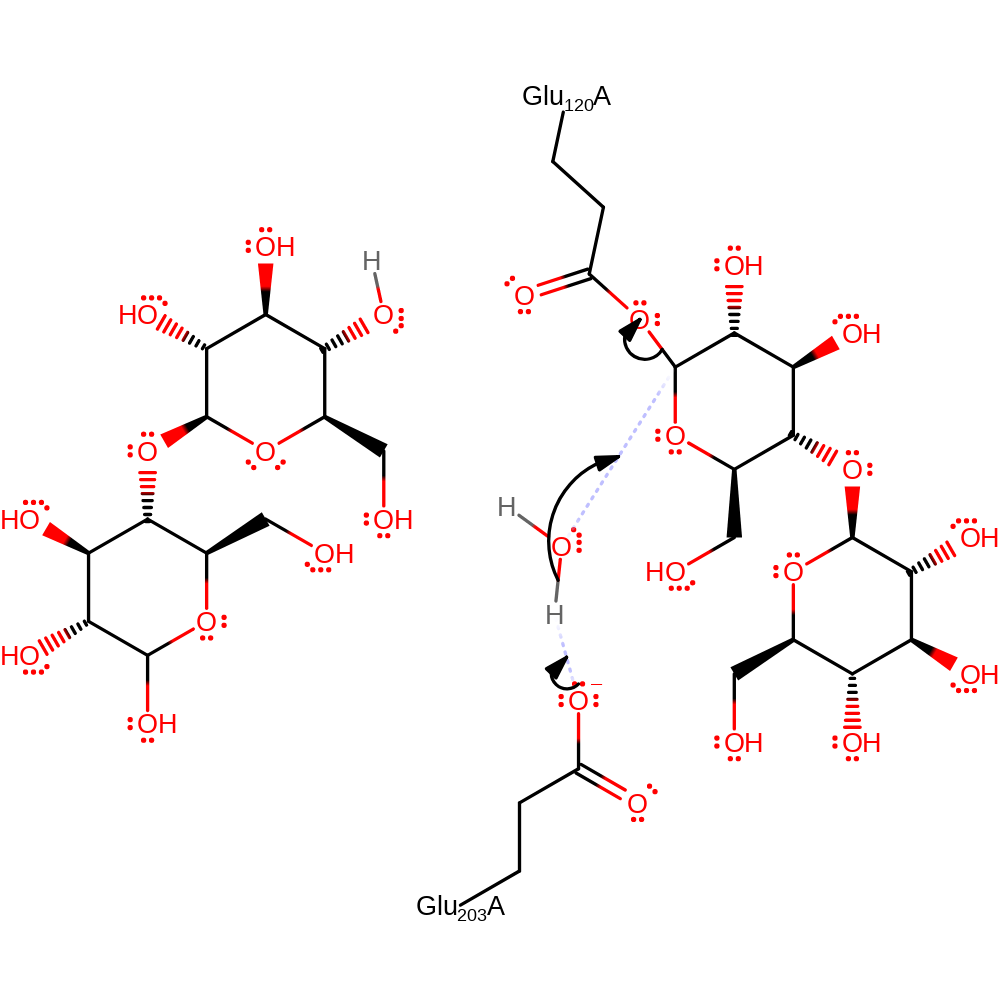
Step 2. Glu203 activates a water molecule which hydrolyses the intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu203A | activator, increase nucleophilicity, proton acceptor |
| Glu120A | nucleofuge |



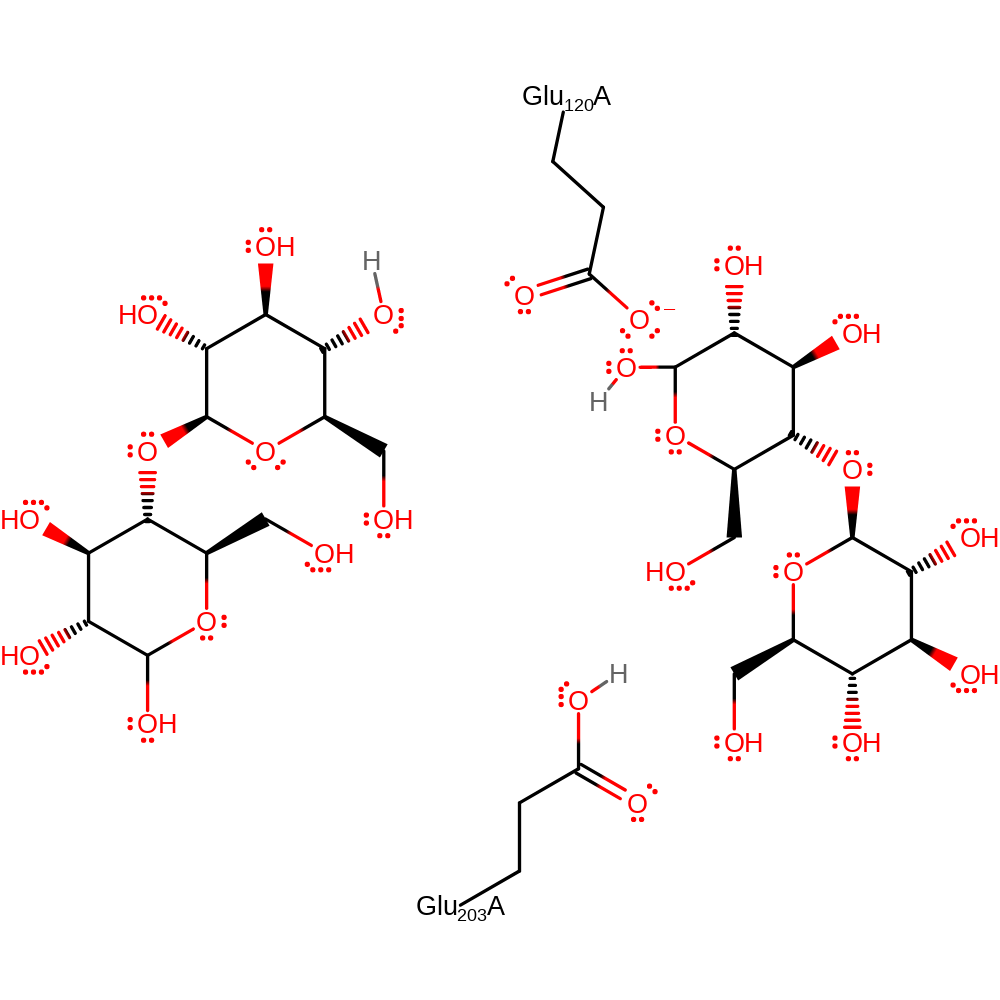 Download:
Download: