L-amino-acid oxidase
L-aminoacid oxidase is a dimeric flavoprotein. it uses a non-covalently bound FAD cofactor in catalysing the stereospecific oxidative deamination of an L-amino acid substrate to an alpha ketoacid, forming ammonia and hydrogen peroxide. The enzyme shows a marked preference for hydrophobic amino acids including phenylalanine, tryptophan, tyrosine and leucine.
Reference Protein and Structure
- Sequence
-
P81382
 (1.4.3.2)
(1.4.3.2)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Calloselasma rhodostoma (Malayan pit viper)

- PDB
-
1f8r
- CRYSTAL STRUCTURE OF L-AMINO ACID OXIDASE FROM CALLOSELASMA RHODOSTOMA COMPLEXED WITH CITRATE
(2.0 Å)



- Catalytic CATH Domains
-
3.90.660.10
 1.10.405.10
1.10.405.10  (see all for 1f8r)
(see all for 1f8r)
- Cofactors
- Fadh2(2-) (1)
Enzyme Mechanism
Introduction
The non-covalently bound FAD is first reduced to FADH2 with oxidation of the amino acid to an imino acid. Non-enzymatic hydrolysis at the imine centre leads to the elimination of ammonia and formation of an alpha-ketoacid. The mechanism is thought to involve a base for the abstraction of a proton from the amino group of the substrate and subsequent transfer of a hydride from the alpha carbon to the N5 of the isoalloxazine ring of FAD to form the imino intermediate.
Catalytic Residues Roles
| UniProt | PDB* (1f8r) | ||
| His241 | His223A | The residue is thought to act as a general base towards the zwitter-ionic form of the substrate, abstracting a proton from the amino group and so activating the substrate to transfer a hydride to the FAD cofactor. | proton acceptor, proton donor |
| Lys344 | Lys326A | The residue acts to relay a proton to a structurally conserved water molecule, which is thought to assist peroxide formation from the flavin-hydroperoxy intermediate. | proton acceptor |
Chemical Components
proton transfer, overall reactant used, aromatic bimolecular nucleophilic addition, hydride transfer, intermediate formation, bimolecular nucleophilic addition, elimination (not covered by the Ingold mechanisms), overall product formed, native state of cofactor regeneratedReferences
- Moustafa IM et al. (2006), J Mol Biol, 364, 991-1002. Crystal Structure of LAAO from Calloselasma rhodostoma with an l-Phenylalanine Substrate: Insights into Structure and Mechanism. DOI:10.1016/j.jmb.2006.09.032. PMID:17046020.
- Pawelek PD et al. (2000), EMBO J, 19, 4204-4215. The structure of L-amino acid oxidase reveals the substrate trajectory into an enantiomerically conserved active site. DOI:10.1093/emboj/19.16.4204. PMID:10944103.

Step 1. His223 acts as a base to deprotonate the amine group of the amino acid substrate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His223A | proton acceptor |
Chemical Components
proton transfer, overall reactant used
Step 2. Once depronated the substrate is activated for hydride transfer from the alpha-C atom to the N5 of FAD, forming the imine intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: aromatic bimolecular nucleophilic addition, hydride transfer, intermediate formation
Step 3. The presence of a structurally conserved water molecule and lysine residue suggest a possible role for Lys326 in activating the water for hydrolysis of the intermediate. The intermediate is subject to nucleophilic attack by the water, while the imine accepts a proton.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His223A | proton donor |
| Lys326A | proton acceptor |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer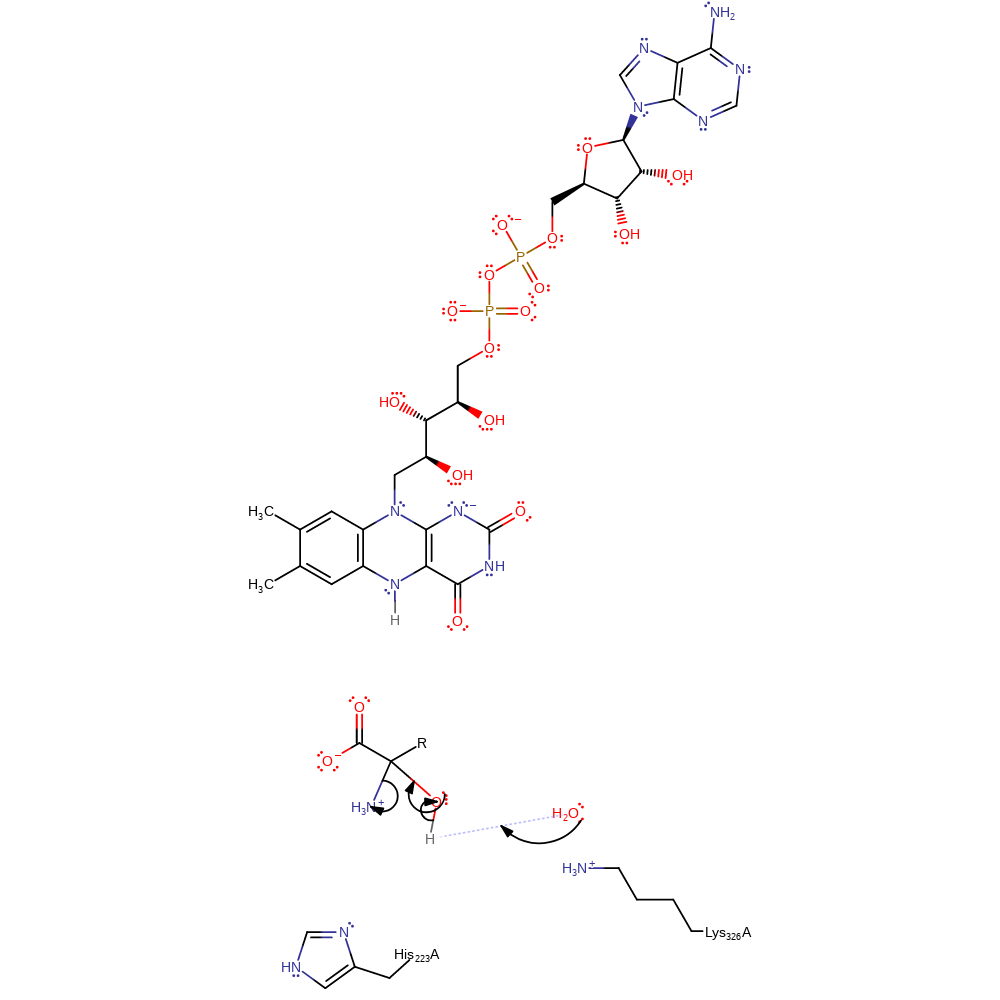
Step 4. In an inferred hydrolysis step there is movement of electrons from the hydroxyl lone pair, causing elimination of ammonia and forming the product. Following hydrolysis there is an oxidative half reaction in which the cofactor FADH- is reoxidised to FAD by molecular oxygen.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|






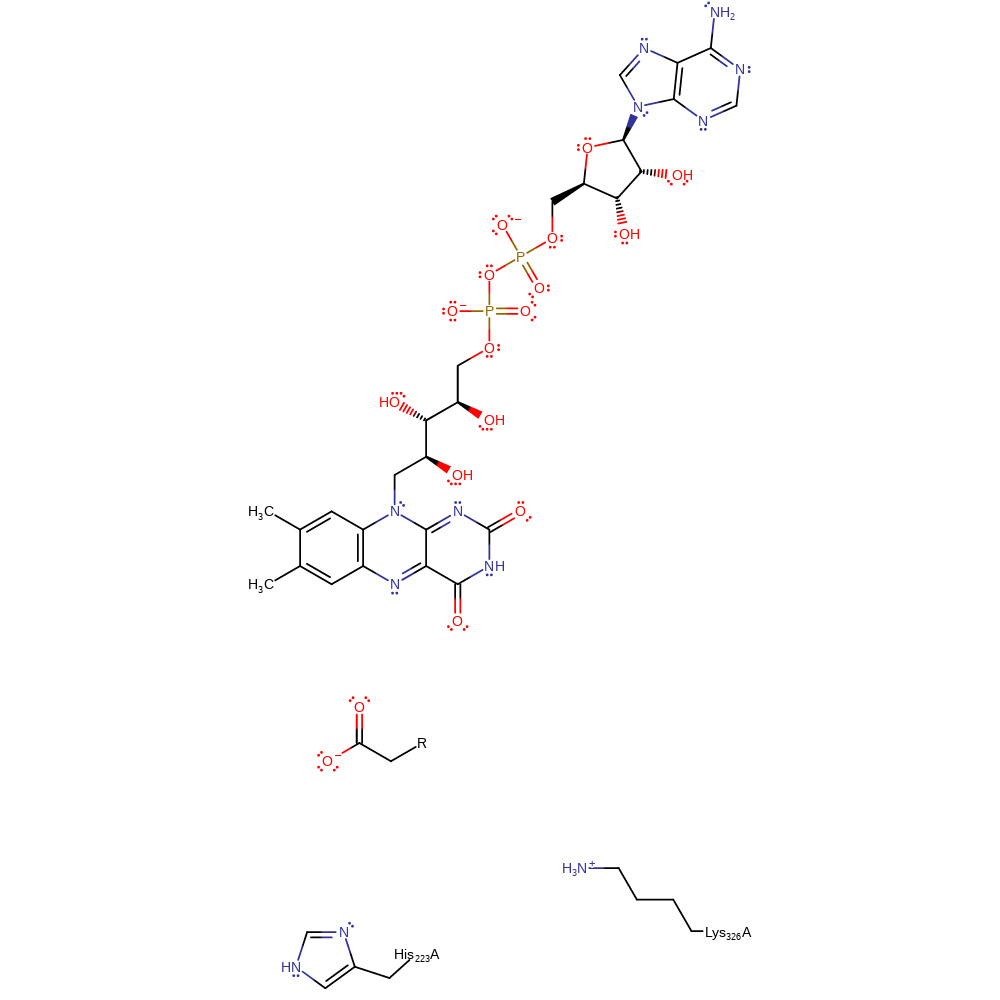 Download:
Download: