Aconitate hydratase
Aconitase (aconitate hydratase) contains an Fe-S cluster within the active site. This can be in the form 4Fe-4S in which case the enzyme functions as an aconitate hydratase, performing the dehydration-rehydration of citrate to isocitrate via cis-aconitate in the second and third steps of the Krebs cycle. When the Fe-S cluster is in the form 3Fe-4S (cytosolic aconitase) the enzyme functions as an Iron Regulatory Protein (IRP) which binds to a conserved sequence (Iron Responsive Element; IRE) in mRNAs encoding proteins that function in the maintenance of iron homeostasis thus regulating the translation of that mRNA. Aconitase occurs in bacteria, plants, animals and fungi in mitochondrial and cytosolic forms.
Reference Protein and Structure
- Sequence
-
P20004
 (4.2.1.3)
(4.2.1.3)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Bos taurus (Cattle)

- PDB
-
1fgh
- COMPLEX WITH 4-HYDROXY-TRANS-ACONITATE
(2.05 Å)



- Catalytic CATH Domains
-
3.30.499.10
 3.20.19.10
3.20.19.10  (see all for 1fgh)
(see all for 1fgh)
- Cofactors
- Water (1), Tetra-mu3-sulfido-tetrairon (1)
Enzyme Reaction (EC:4.2.1.3)
Enzyme Mechanism
Introduction
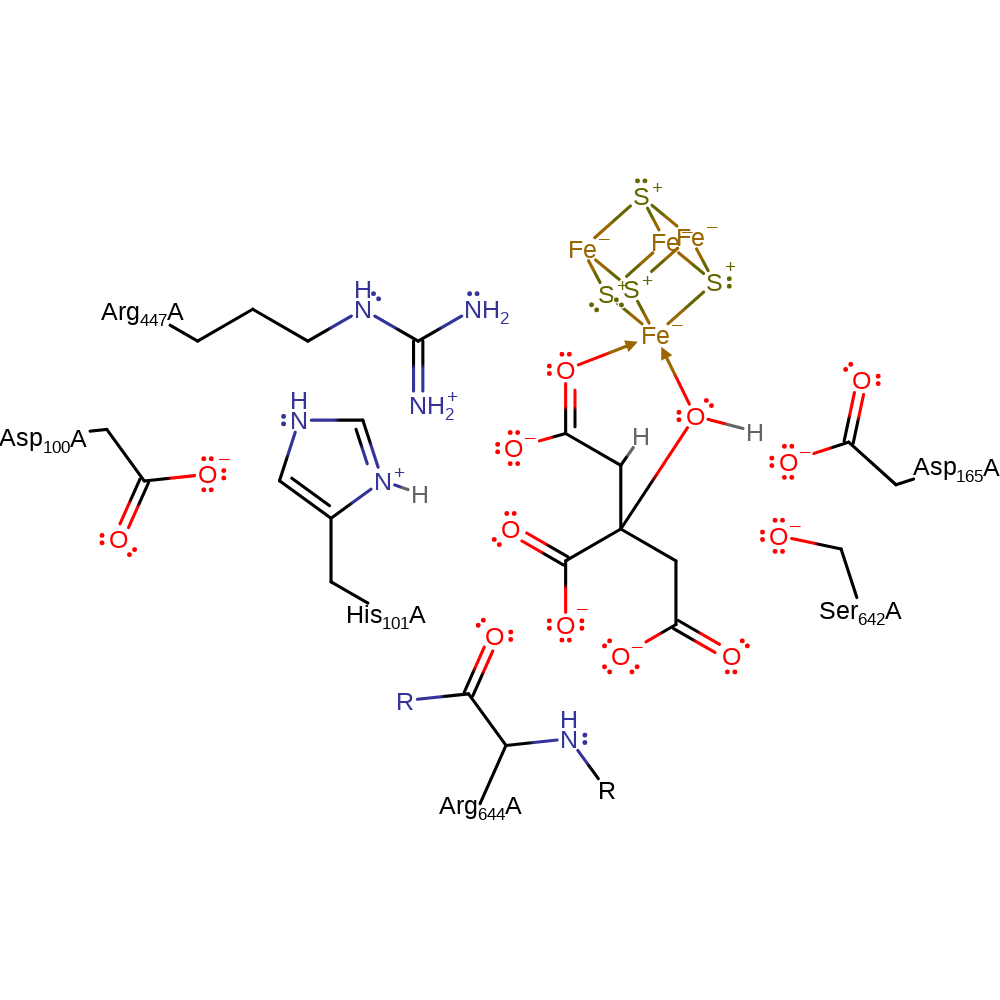
Ser642, the general base is thought to be deprotonated when the isocitrate substrate binds to the Fe centre and Fe-OH2 is formed in the coordination sphere. Serine 642 abstracts the proton from the beta-carbon of isocitrate to form cis-aconitate. Conjugate elimination of the hydroxyl group and cleavage of the alpha C-OH bond results in the deprotonation of His101. The His101-Asp100 pair activate the Fe coordinated water molecule to provide a hydroxide to cis-aconitate. Ser642 then protonates the alpha carbon of this intermediate to form citrate.
Catalytic Residues Roles
| UniProt | PDB* (1fgh) | ||
| His128 | His101A | The residue acts as a general acid to the eliminated hydroxyl from isocitrate. Hydrogen bonding with Asp100 stabilises the residue's positive charge. | proton acceptor, electrostatic stabiliser, proton donor |
| Asp127 | Asp100A | The residue stabilises the positive charge on His101, facilitating the the role of His101 as a general acid. | electrostatic stabiliser, increase acidity |
| Arg671 (main-N) | Arg644A (main-N) | The residue's backbone amide stabilises the alkoxide transition state in the elimination of OH from the isocitrate substrate. | electrostatic stabiliser |
| Arg474 | Arg447A | The interaction between the residue's gaunidinium group and one of the substrate carboxylates enhances the active site specificity for substrates structure and stereochemistry. | electrostatic stabiliser |
| Ser669 | Ser642A | The residue undergoes rapid proton exchange on binding of the substrate and water to the Fe metal centre. In the ionised form the residue then acts as a general base towards isocitrate, abstracting the beta-carbon proton. After elimination and reinsertion of water, Ser642 acts as a general acid to the alpha carbon of the cis-aconitate intermediate, forming the citrate product. | proton acceptor, proton donor |
| Asp192 | Asp165A | The residue hydrogen bonds to both the hydroxyl group of the isocitrate and the coordinated hydroxide. This allows Asp165 to orientate the substrate correctly within the metal coordination sphere for catalysis. | electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular elimination, overall reactant used, bimolecular nucleophilic addition, overall product formed, native state of enzyme regeneratedReferences
- Brown NM et al. (1998), Proc Natl Acad Sci U S A, 95, 15235-15240. Novel role of phosphorylation in Fe-S cluster stability revealed by phosphomimetic mutations at Ser-138 of iron regulatory protein 1. DOI:10.1073/pnas.95.26.15235. PMID:9860952.
- Lloyd SJ et al. (1999), Protein Sci, 8, 2655-2662. The mechanism of aconitase: 1.8 A resolution crystal structure of the S642a:citrate complex. DOI:10.1110/ps.8.12.2655. PMID:10631981.
- Irvin SD et al. (1998), J Mol Evol, 46, 401-408. A unique fungal lysine biosynthesis enzyme shares a common ancestor with tricarboxylic acid cycle and leucine biosynthetic enzymes found in diverse organisms. DOI:10.1007/pl00006319. PMID:9541534.
- Lauble H et al. (1994), J Mol Biol, 237, 437-451. Crystal structures of aconitase with trans-aconitate and nitrocitrate bound. DOI:10.1006/jmbi.1994.1246. PMID:8151704.
- Zheng L et al. (1992), J Biol Chem, 267, 7895-7903. Mutational analysis of active site residues in pig heart aconitase. PMID:1313811.
- Lauble H et al. (1992), Biochemistry, 31, 2735-2748. Crystal structures of aconitase with isocitrate and nitroisocitrate bound. DOI:10.1021/bi00125a014. PMID:1547214.
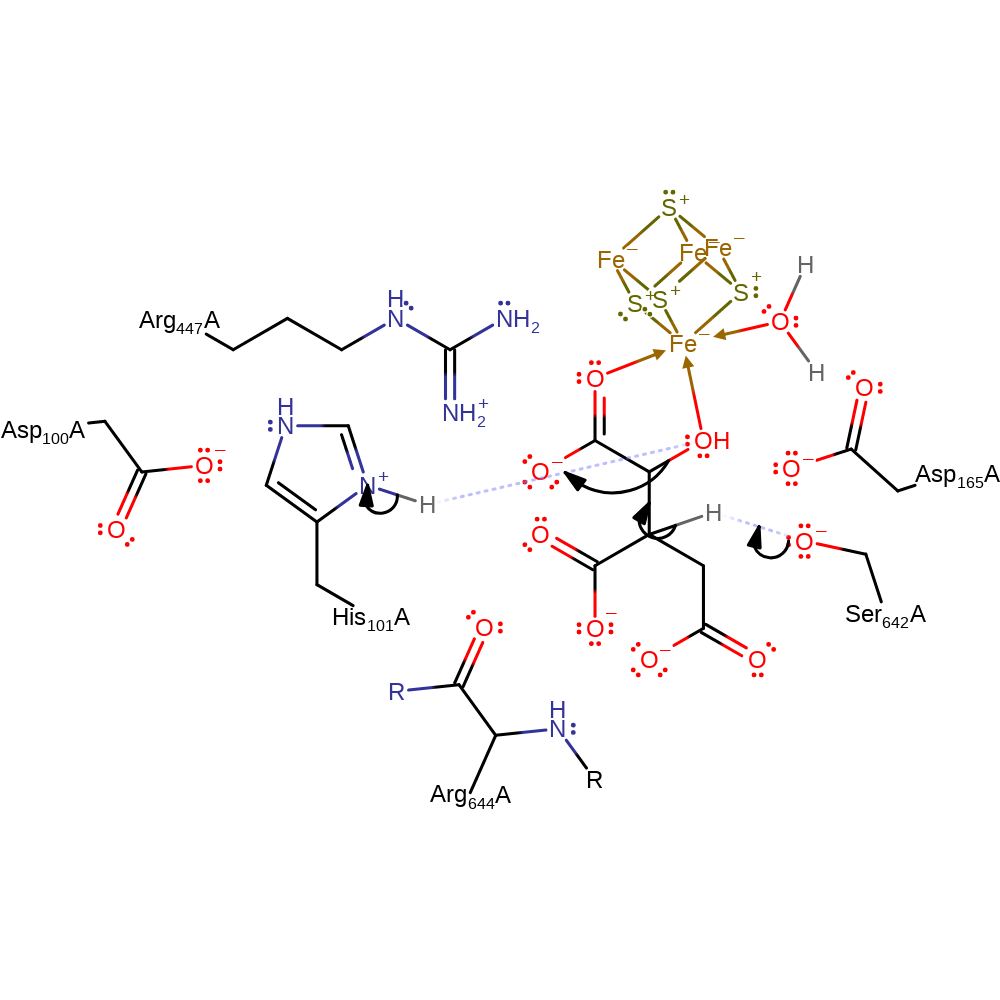
Step 1. Ser642 acts as a general base and deprotonates the beta-carbon of isocitrate. This causes the elimination of the alpha carbon hydroxyl upon protonation by His101 and a cis-aconitate intermediate is formed.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His101A | electrostatic stabiliser |
| Asp100A | electrostatic stabiliser |
| Asp165A | electrostatic stabiliser |
| Arg447A | electrostatic stabiliser |
| Arg644A (main-N) | electrostatic stabiliser |
| Asp100A | increase acidity |
| His101A | proton donor |
| Ser642A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular elimination, overall reactant used
Step 2. His 101 activates the Fe coordinated water which performs a nucleophilic on the beta-carbon. Simultaneously Ser642 acts as a general acid donating a proton to the alpha-carbon, forming the citrate product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp100A | electrostatic stabiliser |
| His101A | electrostatic stabiliser |
| Asp165A | electrostatic stabiliser |
| Arg447A | electrostatic stabiliser |
| Arg644A (main-N) | electrostatic stabiliser |
| Ser642A | proton donor |
| His101A | proton acceptor |


 Download:
Download: