3-dehydroquinate dehydratase (type II)
3-Dehydroquinate dehydratase catalyses the dehydration of 3-dehydroquinate to 3-dehydroshikimate. This reaction is part of both the biosynthetic shikimate pathway for organic compound synthesis and the catabolic quinate pathway using quinate as an energy and carbon source. These pathways are both absent in animals. Type I and type II dehydroquinases exist, with completely different mechanisms and sequences, and this enzyme in question is part of the latter class. The existence of two types allows the targeting of drugs for type II to be developed.
Reference Protein and Structure
- Sequence
-
P15474
 (4.2.1.10)
(4.2.1.10)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Streptomyces coelicolor A3(2) (Bacteria)

- PDB
-
1gu1
- Crystal structure of type II dehydroquinase from Streptomyces coelicolor complexed with 2,3-anhydro-quinic acid
(1.8 Å)



- Catalytic CATH Domains
-
3.40.50.9100
 (see all for 1gu1)
(see all for 1gu1)
- Cofactors
- Water (1)
Enzyme Reaction (EC:4.2.1.10)
Enzyme Mechanism
Introduction
3-Dehydroquinate dehydratase catalyses a trans-dehydration via an enolate intermediate. Tyr 28 acts as a general base catalyst in abstracting a proton from C2, and is assisted by having its pKa lowered by Arg 23 and Arg 113. A water molecule activated by Asn 16 is deprotonated, and protonated by Asn 16 acting as a general acid catalyst. A water molecule is then activated by Asn 16 acting as a general base catalyst to deprotonate C1 and cause loss of a water molecule from the C3 hydroxyl position by His 106 acting as a general acid catalyst. It is activated by Glu 104 through a hydrogen bonding arrangement. Release of the product follows.
Catalytic Residues Roles
| UniProt | PDB* (1gu1) | ||
| Glu105 | Glu104A | Activates His 106. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Asn17 | Asn16A | Acts as a general acid/base catalyst to activate water in proton donation and abstraction. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Ala83 (main-N), Pro16 (main-C), Asn17 | Ala82A (main-N), Pro15A (main-C), Asn16A | Hold a water molecule in place so that it can act as an electrostatic stabiliser. | activator, hydrogen bond donor, steric role |
| Tyr29 | Tyr28A | Acts as a general base catalyst in deprotonation of the substrate at the C2 position. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| His107 | His106A | Acts as a general acid catalyst in donation of a proton to the substrate to form water in a condensation reaction. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, proton relay |
| Asn80 | Asn79A | Helps stabilise the reaction intermediates. | hydrogen bond acceptor, steric role |
| Arg24, Arg114 | Arg23A, Arg113A | Activates Tyr 28 by lowering its pKa. | attractive charge-charge interaction, activator, electrostatic stabiliser, polar interaction |
Chemical Components
proton transfer, assisted keto-enol tautomerisation, intermediate formation, overall reactant used, proton relay, bimolecular elimination, overall product formed, intermediate terminated, intermediate collapse, dehydration, native state of enzyme regenerated, inferred reaction stepReferences
- Roszak AW et al. (2002), Structure, 10, 493-503. The Structure and Mechanism of the Type II Dehydroquinase from Streptomyces coelicolor. DOI:10.1016/s0969-2126(02)00747-5. PMID:11937054.
- Reiling S et al. (2014), Acta Crystallogr F Struct Biol Commun, 70, 1485-1491. Structure of type II dehydroquinase fromPseudomonas aeruginosa. DOI:10.1107/s2053230x14020214. PMID:25372814.
- Chu W et al. (2013), Mol Simul, 39, 137-144. Molecular dynamics (MD) simulations and binding free energy calculation studies between inhibitors and type II dehydroquinase (DHQ2). DOI:10.1080/08927022.2012.708416.
- Payne RJ et al. (2007), ChemMedChem, 2, 1010-1013. Design, Synthesis, and Structural Studies on Potent Biaryl Inhibitors of Type II Dehydroquinases. DOI:10.1002/cmdc.200700062. PMID:17487901.
- González-Bello C et al. (2007), Med Res Rev, 27, 177-208. Progress in type II dehydroquinase inhibitors: From concept to practice. DOI:10.1002/med.20076. PMID:17004270.
- Robinson DA et al. (2006), J Med Chem, 49, 1282-1290. Crystal Structures ofHelicobacterpyloriType II Dehydroquinase Inhibitor Complexes: New Directions for Inhibitor Design. DOI:10.1021/jm0505361. PMID:16480265.
- Toscano MD et al. (2005), Org Biomol Chem, 3, 3102-3104. Rational design of new bifunctional inhibitors of type II dehydroquinase. DOI:10.1039/b507156a. PMID:16106291.
- Frederickson M et al. (2004), Org Biomol Chem, 2, 1592-1596. (1R,4S,5R)-3-Fluoro-1,4,5-trihydroxy-2-cyclohexene-1-carboxylic acid: the fluoro analogue of the enolate intermediate in the reaction catalyzed by type II dehydroquinases. DOI:10.1039/b404535a. PMID:15162210.
- Maes D et al. (2004), Acta Crystallogr D Biol Crystallogr, 60, 463-471. Structural study of the type II 3-dehydroquinate dehydratase fromActinobacillus pleuropneumoniae. DOI:10.1107/s090744490302969x. PMID:14993670.
- Lee BI et al. (2003), Proteins, 51, 616-617. Crystal structure of the type II 3-dehydroquinase from Helicobacter pylori. DOI:10.1002/prot.10415. PMID:12784220.
- Gourley DG et al. (1999), Nat Struct Biol, 6, 521-525. The two types of 3-dehydroquinase have distinct structures but catalyze the same overall reaction. DOI:10.1038/9287. PMID:10360352.
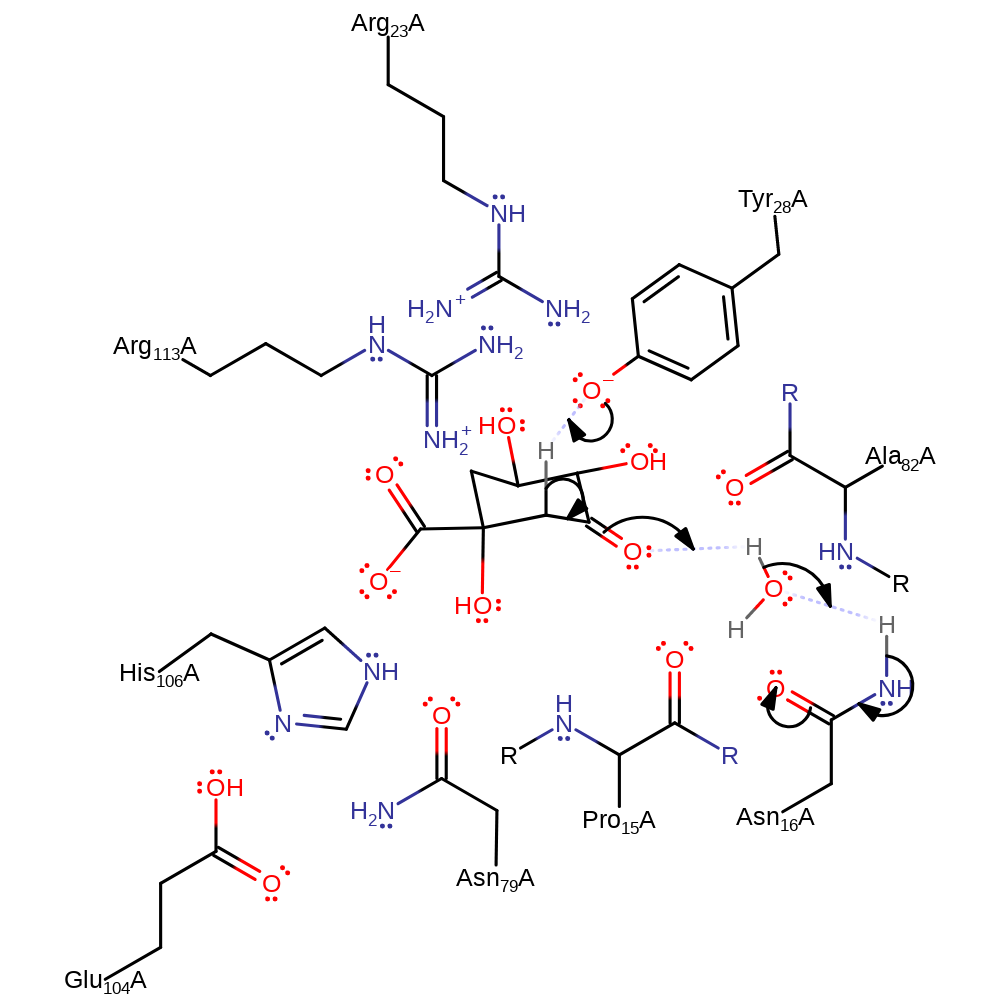
Step 1. Tyr28 deprotonates the substrate, forming a double bond and causing the ketone group to reduce and deprotonate water, which in turn deprotonates Asn16.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn79A | hydrogen bond acceptor |
| Arg113A | attractive charge-charge interaction, electrostatic stabiliser |
| His106A | hydrogen bond acceptor, hydrogen bond donor |
| Pro15A (main-C) | hydrogen bond acceptor, activator, steric role |
| Asn16A | hydrogen bond donor |
| Arg23A | attractive charge-charge interaction, electrostatic stabiliser |
| Tyr28A | hydrogen bond acceptor |
| Ala82A (main-N) | hydrogen bond donor, activator, steric role |
| Glu104A | hydrogen bond donor |
| Asn16A | proton donor |
| Tyr28A | proton acceptor |
Chemical Components
proton transfer, assisted keto-enol tautomerisation, intermediate formation, overall reactant used, proton relay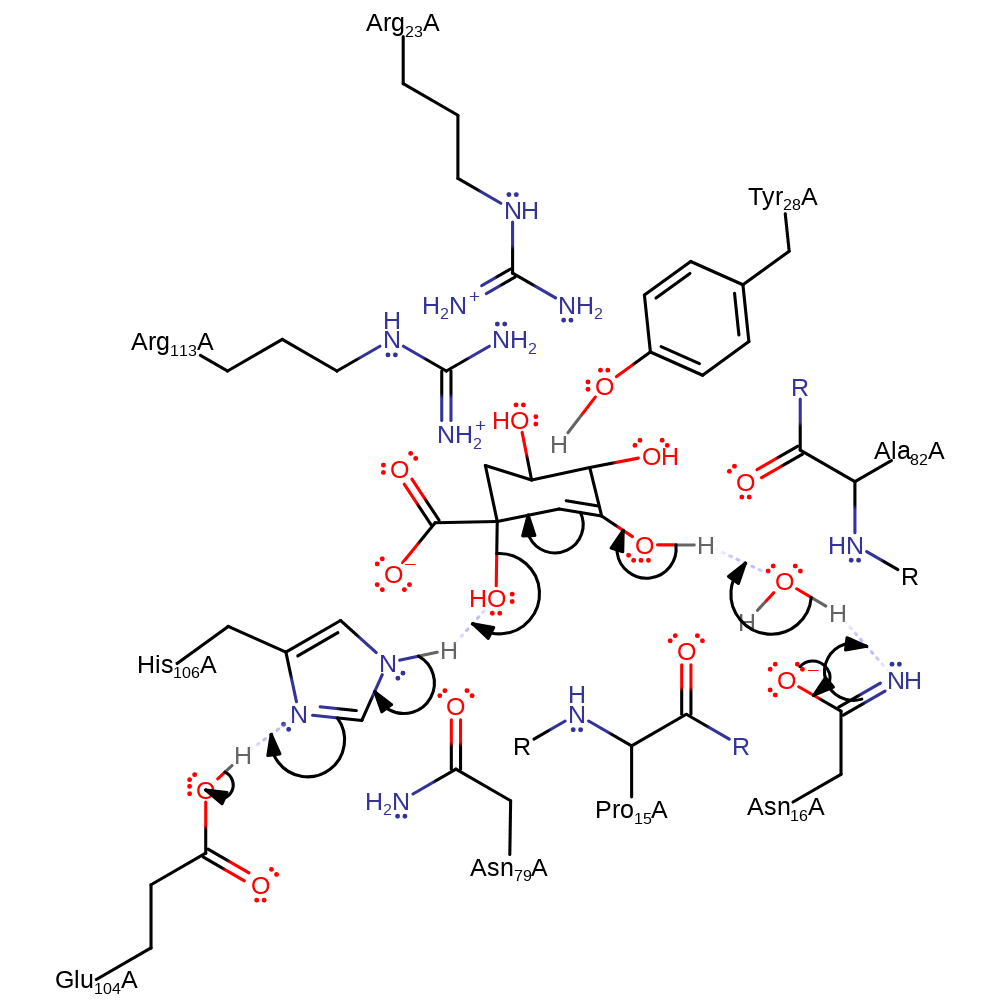
Step 2. Asn16 deprotonates water, which deprotonates the C1-hydroxy, causing a double bond rearrangement that eliminates water with deprotonation of His106, which in turn deprotonates Glu104.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn79A | hydrogen bond acceptor, steric role |
| Arg113A | polar interaction, electrostatic stabiliser |
| His106A | hydrogen bond acceptor, hydrogen bond donor |
| Pro15A (main-C) | hydrogen bond acceptor, steric role, activator |
| Asn16A | hydrogen bond acceptor |
| Arg23A | polar interaction, electrostatic stabiliser |
| Ala82A (main-N) | hydrogen bond donor, steric role, activator |
| Glu104A | hydrogen bond donor |
| Asn16A | proton acceptor |
| His106A | proton donor |
| Glu104A | proton donor |
| His106A | proton acceptor, proton relay |
Chemical Components
ingold: bimolecular elimination, proton transfer, overall product formed, intermediate terminated, intermediate collapse, dehydration, proton relay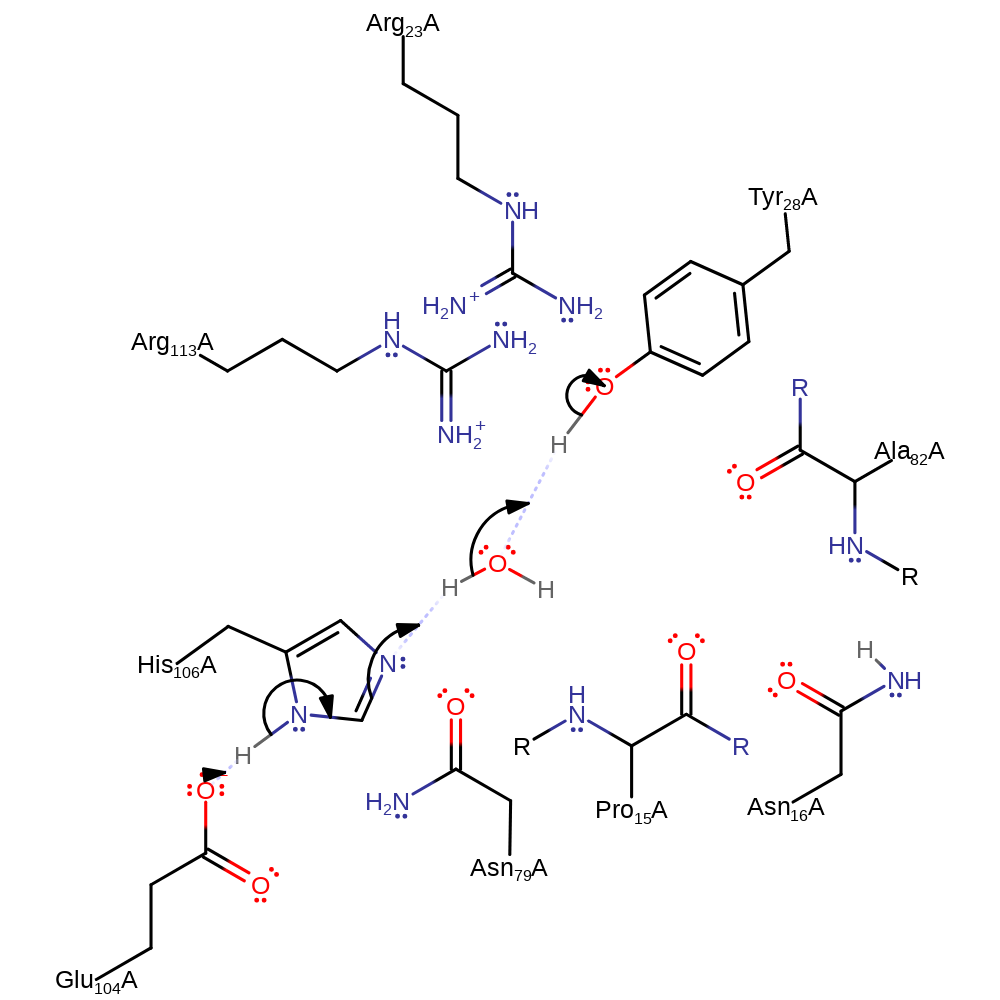
Step 3. Glu104 deprotonates His106, which deprotonates water, which in turn deprotonates Tyr28 in an inferred step.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn79A | hydrogen bond acceptor |
| Arg113A | polar interaction, activator |
| His106A | hydrogen bond acceptor, hydrogen bond donor |
| Pro15A (main-C) | hydrogen bond acceptor, steric role, activator |
| Asn16A | hydrogen bond acceptor, hydrogen bond donor |
| Arg23A | polar interaction, activator |
| Tyr28A | hydrogen bond donor |
| Ala82A (main-N) | hydrogen bond donor |
| Glu104A | hydrogen bond acceptor |
| His106A | proton acceptor |
| Glu104A | proton acceptor |
| His106A | proton donor |
| Tyr28A | proton donor |
| His106A | proton relay |



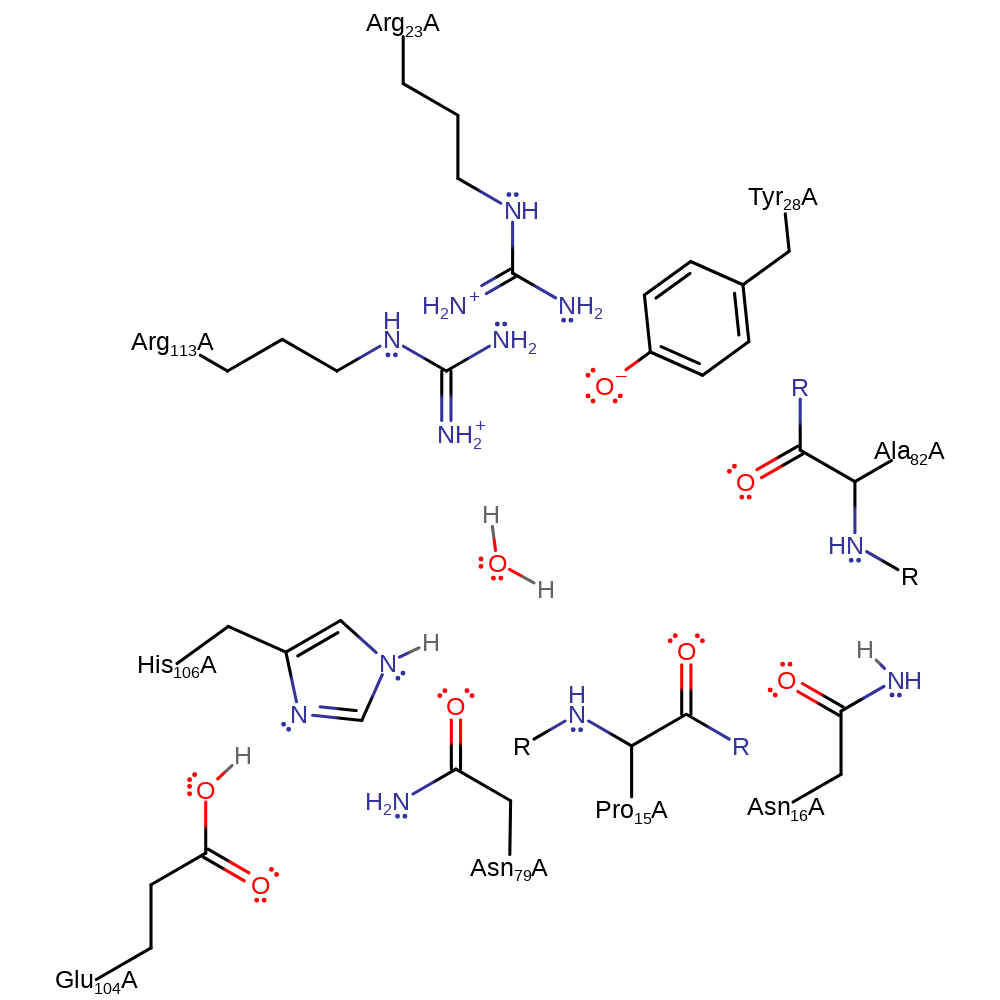 Download:
Download: