Carbonate dehydratase (gamma class)
Gamma-class carbonic anhydrase (Cam) from Methanosarcina thermophila catalyses the reversible hydration of carbon dioxide to bicarbonate. There are three distinct classes of carbonic anhydrases (alpha, beta and gamma). In M. thermophila, Cam activity increases when the substrate for growth is acetate, suggesting the involvement of this enzyme in acetate catabolism.
Reference Protein and Structure
- Sequence
-
P40881
 (4.2.1.1)
(4.2.1.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Methanosarcina thermophila TM-1 (Archaea)

- PDB
-
1qrg
- A CLOSER LOOK AND THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTALLOGRAPHIC STUDIES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA
(1.72 Å)



- Catalytic CATH Domains
-
2.160.10.10
 (see all for 1qrg)
(see all for 1qrg)
- Cofactors
- Cobalt(3+) (1)
Enzyme Reaction (EC:4.2.1.1)
Enzyme Mechanism
Introduction
Glu 62 deprotonates a water molecule liganded to Co. This proton is then abstracted by Glu 84 (and ultimately shuttled out to buffer.) This hydroxyl ligand then abstracts a proton from the adjacent liganded water molecule. The newly formed hydroxide ligand is stabilised by hydrogen bonding to Gln 75. CO2 is polarised by Asn 202 to make the carbon more attractive for nucleophilic attack. The hydroxide ligand nucleophilically attacks the carbon atom of CO2, resulting in a bound HCO3-. An oxygen atom of HCO3- displaces a liganded water molecule from Coto form a bidentate HCO3- ligand. HCO3- undergoes a bidentate transition state where the proton either rotates (Lindskog) or transfers (Lipscomb) to the non-metal-bound oxygen of HCO3-. Glu 62 destabilises HCO3- by hydrogen bonding to it. A water molecule replaces one of the bound oxygens of the HCO3- ligand. A second water molecule replaces the other bound oxygen atom of HCO3-, displacing it completely from Co.
Catalytic Residues Roles
| UniProt | PDB* (1qrg) | ||
| Glu96 | Glu62A | Acts as a base by deprotonating a water molecule liganded to Zn. Destabilises HCO3- ligand by hydrogen bonding to it, aiding its detachment from Zn. | proton acceptor, electrostatic destabiliser, electrostatic stabiliser, proton donor |
| Glu118 | Glu84A | Abstracts a proton from Glu 62 and shuttles it up to the surface buffer. | proton acceptor |
| His115, His156, His151 | His81A, His122A, His117A(AA) | Bind the metal ion. | metal ligand |
| Asn236 | Asn202A(AA) | Polarises CO2, assisting the nucleophilic attack of HO-. | increase electrophilicity, electrostatic stabiliser |
| Gln109 | Gln75A(AA) | Stabilises the hydroxide ligand, readying it for nucleophilic attack on CO2. | electrostatic stabiliser |
Chemical Components
proton transfer, proton relay, bimolecular nucleophilic addition, overall reactant used, overall product formed, decoordination from a metal ion, coordination to a metal ion, native state of cofactor regeneratedReferences
- Zimmerman SA et al. (2006), Biochemistry, 45, 5149-5157. Proposal for a Hydrogen Bond Network in the Active Site of the Prototypic γ-Class Carbonic Anhydrase†. DOI:10.1021/bi052507y. PMID:16618104.
- Ghiasi M et al. (2017), Comput Theor Chem, 1109, 42-57. Activation modelling of β- and γ-class of carbonic anhydrase with amines and amino acids: Proton transfer process within the active site from thermodynamic point of view. DOI:10.1016/j.comptc.2017.03.041.
- Ferry JG (2010), Biochim Biophys Acta, 1804, 374-381. The gamma class of carbonic anhydrases. DOI:10.1016/j.bbapap.2009.08.026. PMID:19747990.
- Iverson TM et al. (2000), Biochemistry, 39, 9222-9231. A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila. PMID:10924115.
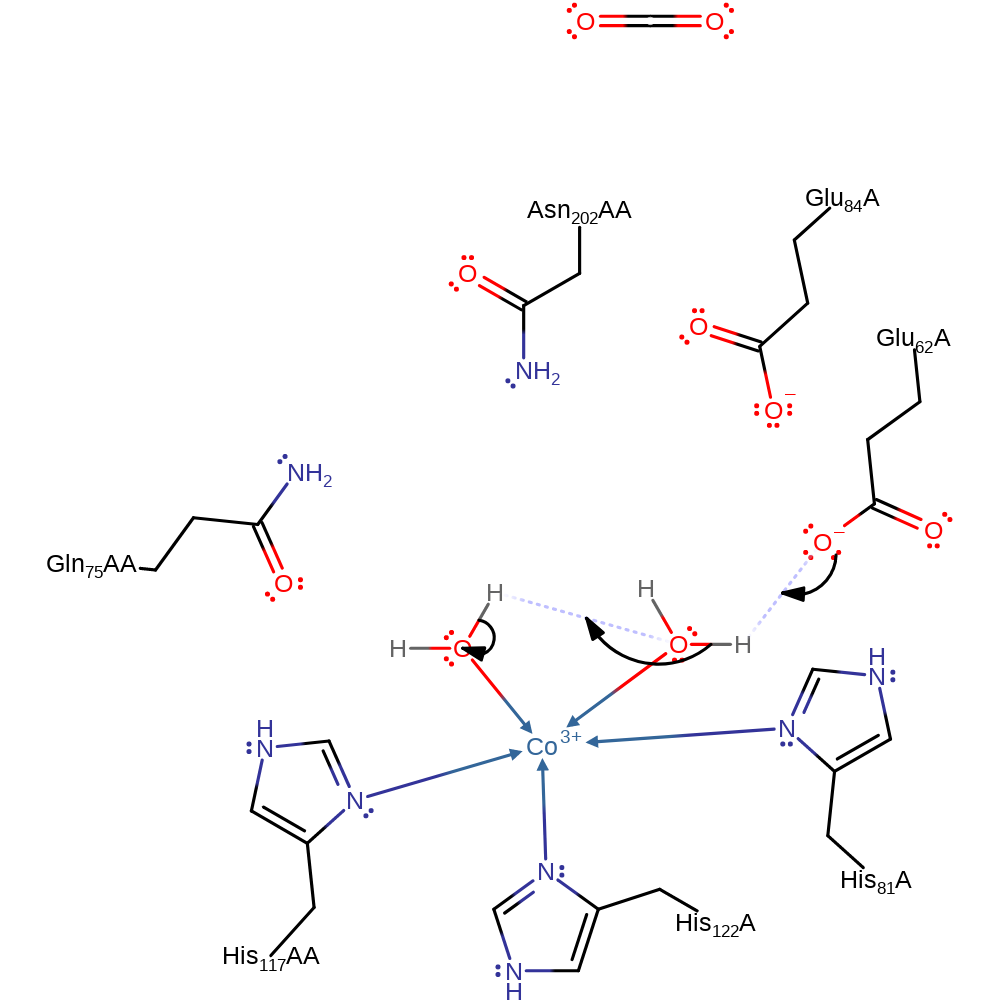
Step 1. Glu 62 deprotonates a water molecule liganded to Co. This hydroxyl ligand then abstracts a proton from the adjacent liganded water molecule. The newly formed hydroxide ligand is stabilised by hydrogen bonding to Gln 75.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln75A(AA) | electrostatic stabiliser |
| Glu62A | electrostatic stabiliser |
| His81A | metal ligand |
| His122A | metal ligand |
| Glu62A | proton acceptor |
Chemical Components
proton transfer, proton relay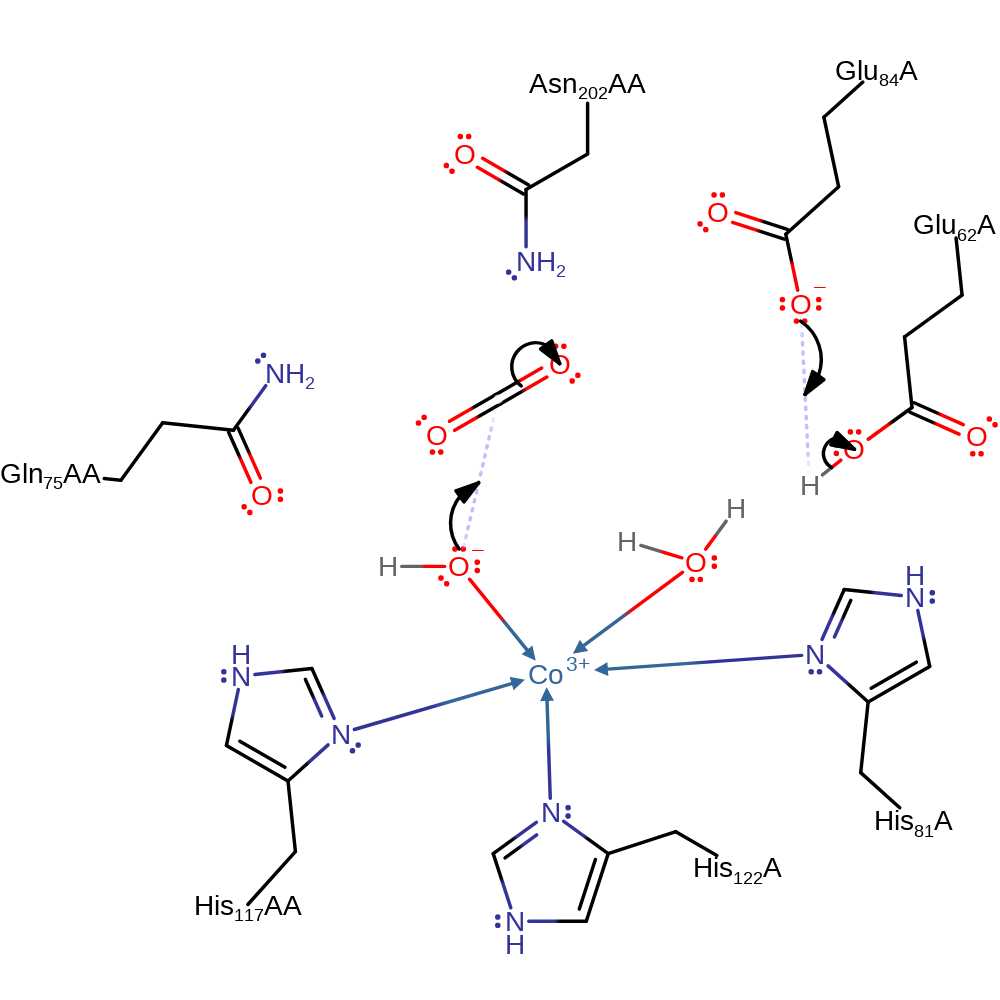
Step 2. The proton bound to Glu62 is then abstracted by Glu84 where it is shuttled into the solution. CO2 is polarised by Asn 202 to make the carbon more attractive for nucleophilic attack. The hydroxide ligand nucleophilically attacks the carbon atom of CO2, resulting in a bound HCO3-.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn202A(AA) | electrostatic stabiliser |
| Gln75A(AA) | electrostatic stabiliser |
| Asn202A(AA) | increase electrophilicity |
| His81A | metal ligand |
| His122A | metal ligand |
| His117A(AA) | metal ligand |
| Glu84A | proton acceptor |
| Glu62A | proton donor |
Chemical Components
ingold: bimolecular nucleophilic addition, overall reactant used, overall product formed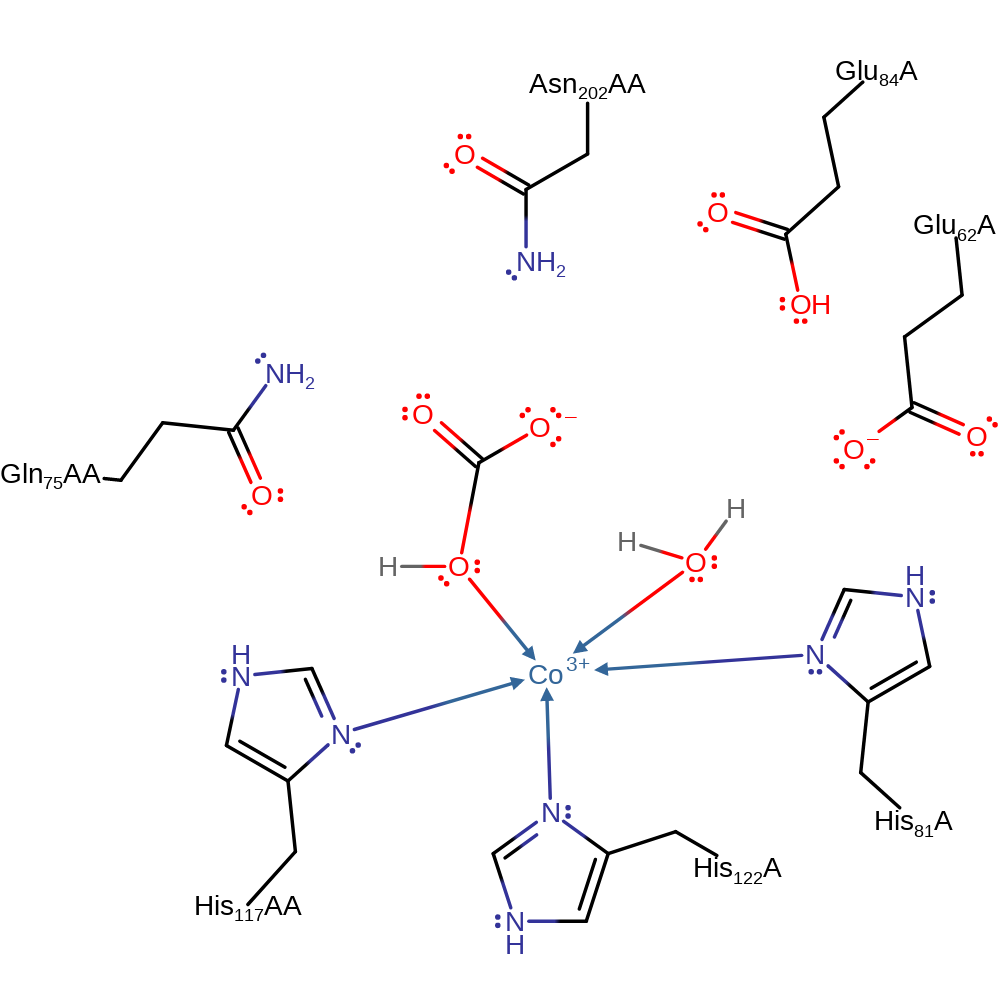
Step 3. An oxygen atom of HCO3- displaces a liganded water molecule from Co to form a bidentate HCO3- ligand.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His81A | metal ligand |
| His122A | metal ligand |
| His117A(AA) | metal ligand |
Chemical Components
decoordination from a metal ion, coordination to a metal ion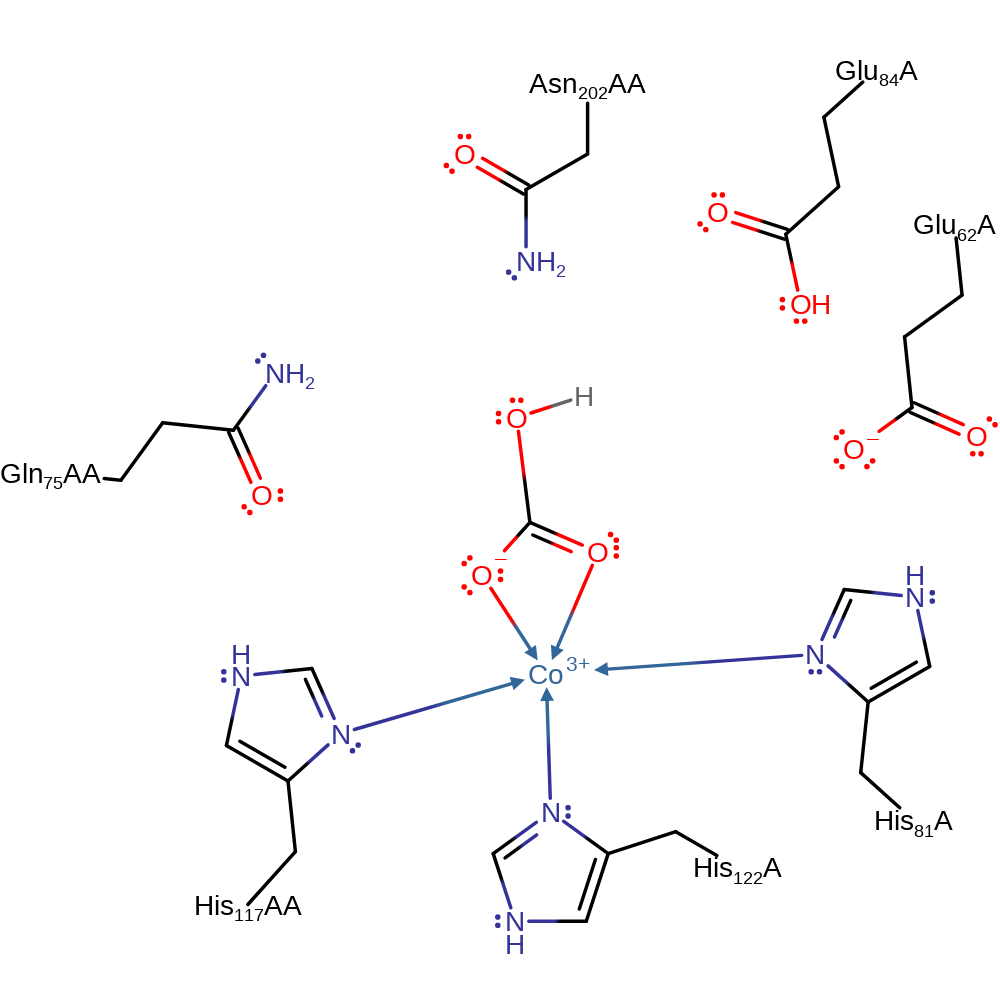
Step 4. Glu 62 destabilises HCO3- by hydrogen bonding to it. A water molecule replaces one of the bound oxygens of the HCO3- ligand.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu62A | electrostatic destabiliser |
| His81A | metal ligand |
| His122A | metal ligand |
| His117A(AA) | metal ligand |
Chemical Components
decoordination from a metal ion, coordination to a metal ion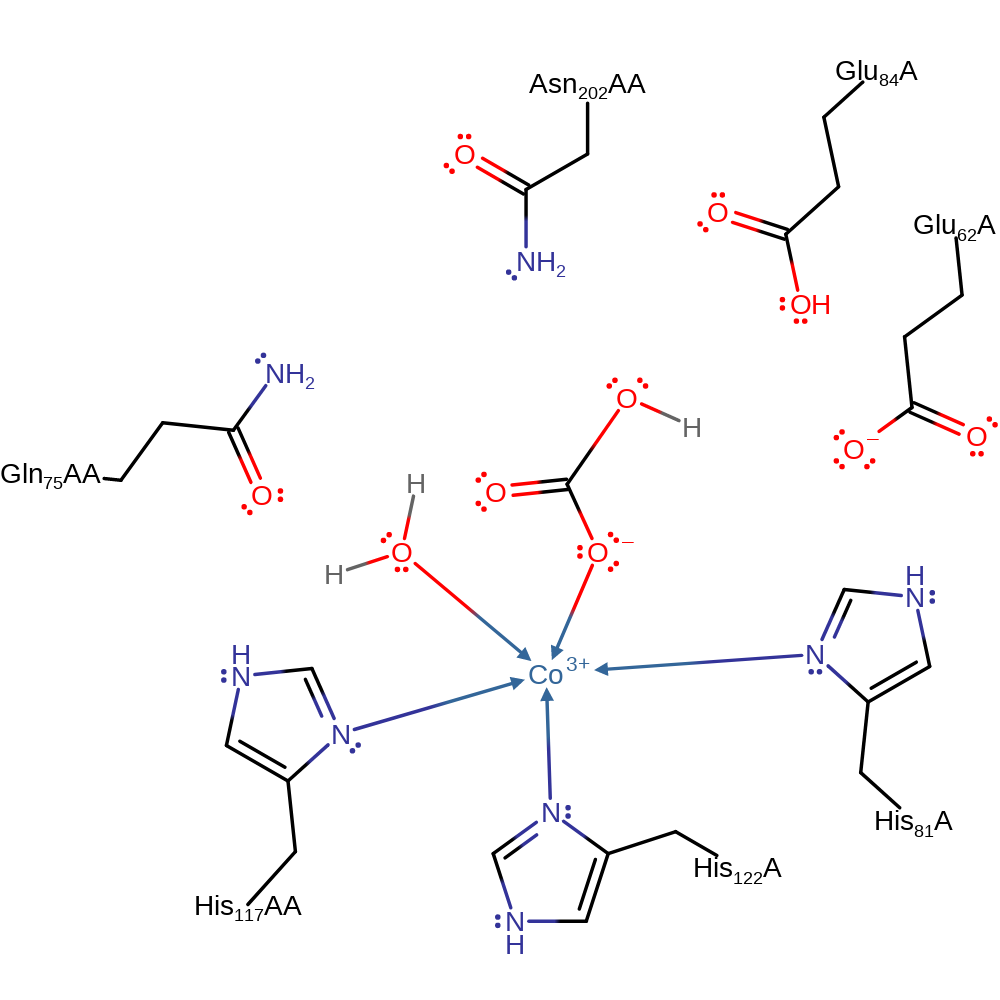
Step 5. A second water molecule replaces the other bound oxygen atom of HCO3-, displacing it completely from Co.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His81A | metal ligand |
| His122A | metal ligand |
| His117A(AA) | metal ligand |




 Download:
Download: