Protein disulfide-isomerase (DsbC)
The disulphide bond isomerase DsbC from E. coli is able to break up incorrectly formed disulphide bonds by transferring electrons to reduce the bond. It displays homology to the thioredoxase family of enzymes.
Reference Protein and Structure
- Sequence
-
P0AEG6

 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1eej
- CRYSTAL STRUCTURE OF THE PROTEIN DISULFIDE BOND ISOMERASE, DSBC, FROM ESCHERICHIA COLI
(1.9 Å)



- Catalytic CATH Domains
-
3.40.30.10
 (see all for 1eej)
(see all for 1eej)
Enzyme Mechanism
Introduction
Asp 95 and Arg 125 increase the nucleophilicity of Cys 98 allowing it to attack the first of the disulphide bonds in a nucleophilic substitution reaction that results in an enzyme bound intermediate with a new thiolate. The newly formed thiolate attacks the second of the disulphide bonds in a nucleophilic substitution reaction that results in a new disulphide bond and a second thiolate. The second newly formed thiolate attacks the disulphide bond between the intermediate and the Cys 98 in a nucleophilic substitution reaction that results in the final new disulphide bond and regenerates Cys 98 in its native state. With complex substrates (i.e. those with more than 2 disulphide bonds) this enzyme uses both catalytic cysteines (Cys98 and Cys101). With simple substrates (i.e. those with only 2 disulphide bonds) this enzyme uses only Cys98. The reaction is shown only for the simple case.
Catalytic Residues Roles
| UniProt | PDB* (1eej) | ||
| Cys118 | Cys98A | Acts as the initial nucleophile catalysing reduction of the disulphide bond in the protein substrate. | nucleofuge, nucleophile, proton acceptor, proton donor |
| Cys121 | Cys101A | Acts as nucleophile to attack a disluphide bond in complex proteins with more than two disulphide bonds. | |
| Asp115, Arg145 | Asp95A, Arg125A | Increases the nucleophilicity of Cys 98. | increase nucleophilicity, proton acceptor, proton donor |
Chemical Components
overall reactant used, intermediate formation, proton transfer, bimolecular nucleophilic substitution, intramolecular nucleophilic substitution, overall product formed, intermediate terminated, native state of enzyme regeneratedReferences
- McCarthy AA et al. (2000), Nat Struct Biol, 7, 196-199. Crystal structure of the protein disulfide bond isomerase, DsbC, from Escherichia coli. DOI:10.1038/73295. PMID:10700276.
- Jiao L et al. (2013), J Struct Biol, 183, 1-10. Crystal structure of the periplasmic disulfide-bond isomerase DsbC from Salmonella enterica serovar Typhimurium and the mechanistic implications. DOI:10.1016/j.jsb.2013.05.013. PMID:23726983.
- Kadokura H et al. (2010), Antioxid Redox Signal, 13, 1231-1246. Mechanisms of oxidative protein folding in the bacterial cell envelope. DOI:10.1089/ars.2010.3187. PMID:20367276.

Step 1. There is nucleophilic attack from Cys 98 to one of the sulfur atoms of the protein disulphide bond. The nucleophilicity of Cys 98 is increased by electrostatic interactions of the Asp and Arg residues. This results in an SN2 reaction, where a new disulphide bond is formed between Cys 98 and the substrate protein, leaving a thiolate group on the other sulfur of the old disulphide bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp95A | increase nucleophilicity |
| Arg125A | increase nucleophilicity |
| Asp95A | proton acceptor |
| Cys98A | proton donor, nucleophile |
Chemical Components
overall reactant used, intermediate formation, proton transfer, ingold: bimolecular nucleophilic substitution
Step 2. The thiolate group of the substrate attacks a sulfur atom of the other disulphide bond in a second SN2 reaction forming a new disulphide bond and a new thiolate group within the substrate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: intramolecular nucleophilic substitution
Step 3. The new thiolate group attacks the substrate sulfur, which has a disulphide bond with Cys 98, in a third SN2 reaction, this forms a a new disulphide bond within the substrate and regenerates Cys 98 in its native state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys98A | proton acceptor |
| Asp95A | proton donor |
| Cys98A | nucleofuge |
Chemical Components
ingold: intramolecular nucleophilic substitution, proton transfer, overall product formed, intermediate terminated, native state of enzyme regeneratedIntroduction
This is the second proposed mechanism for the repair of a misoxidized cysteine pair, where the enzyme acts as a reductase rather than as an isomerase. In common with the other mechanism there is nucleophillic attack from Cys 98, promoted by Asp 95 and Arg125, which breaks the disulphide bond of the substrate protein and forms an intermediate disulphide bond between Cys 98 and the substrate. A thiolate is formed on the other sulfur of the substrate. Nucleophilic attack from Cys 101 breaks the disulphide bond between the substrate and Cys 98 in a second SN2 reaction and a new disulphide bond is formed between the two enzyme cysteine residues. This leaves both the sulfur atoms of the substrate reduced.
Catalytic Residues Roles
| UniProt | PDB* (1eej) | ||
| Cys118 | Cys98A | Acts as a nucleophile to break the disulphide bond of the misoxidized protein. | electrofuge, electrophile, nucleophile, proton donor |
| Cys121 | Cys101A | Acts as a nucleophile to break the disulphide bond between Cys 98 and the substrate, forming a disulphide bond between the two enzyme cysteines. | nucleophile, proton donor |
| Asp115, Arg145 | Asp95A, Arg125A | Increase the nucleophilicity of Cys 98. | increase nucleophilicity, proton acceptor, proton donor |
Chemical Components
bimolecular nucleophilic substitution, proton transfer, intermediate formation, overall reactant used, native state of enzyme is not regenerated, overall product formed, intermediate terminatedReferences
- Jiao L et al. (2013), J Struct Biol, 183, 1-10. Crystal structure of the periplasmic disulfide-bond isomerase DsbC from Salmonella enterica serovar Typhimurium and the mechanistic implications. DOI:10.1016/j.jsb.2013.05.013. PMID:23726983.
- Kadokura H et al. (2010), Antioxid Redox Signal, 13, 1231-1246. Mechanisms of oxidative protein folding in the bacterial cell envelope. DOI:10.1089/ars.2010.3187. PMID:20367276.

Step 1. There is nucleophilic attack from Cys 98 to one of the sulfur atoms of the protein disulphide bond. The nucleophilicity of Cys 98 is increased by electrostatic interactions of the Asp and Arg residues. This results in an SN2 reaction, where a new disulphide bond is formed between Cys 98 and the substrate protein, leaving a thiolate group on the other sulfur of the old disulphide bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp95A | increase nucleophilicity |
| Arg125A | increase nucleophilicity |
| Asp95A | proton acceptor |
| Cys98A | proton donor, nucleophile |
Chemical Components
ingold: bimolecular nucleophilic substitution, proton transfer, intermediate formation, overall reactant used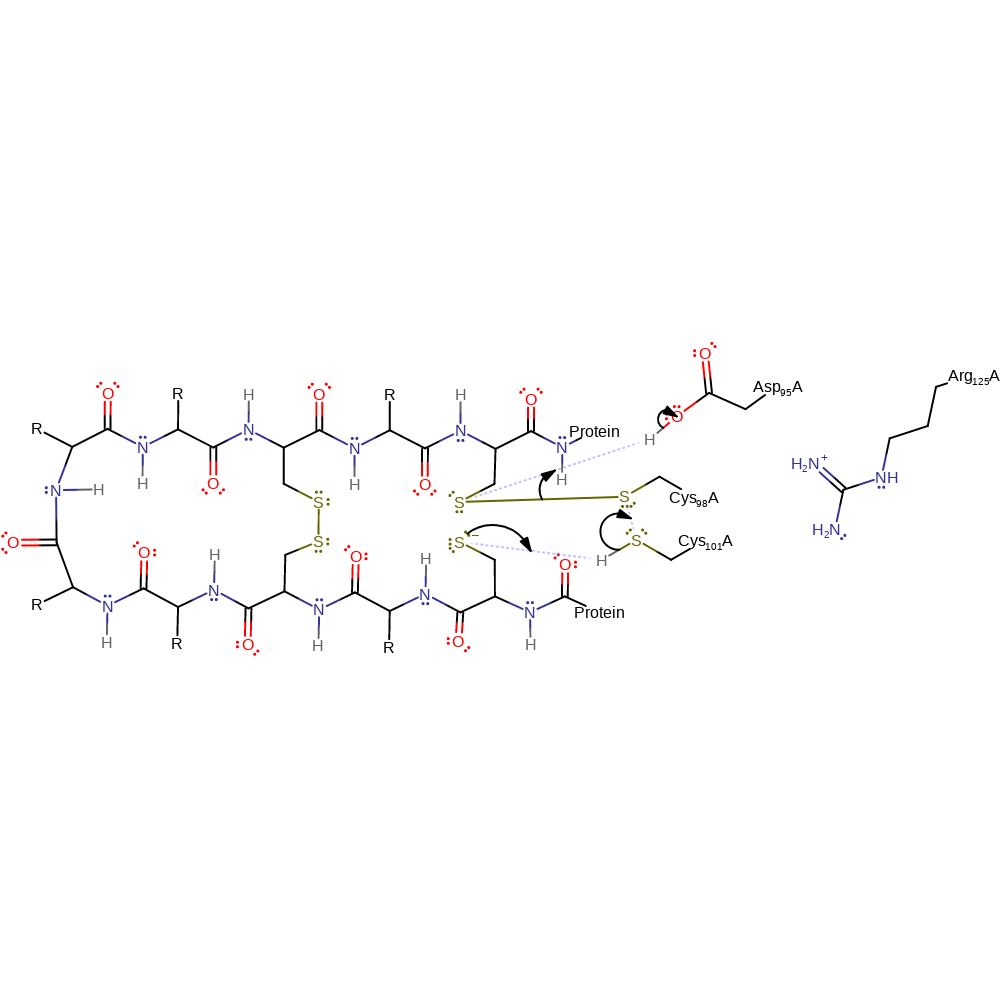
Step 2. There is a second SN2 reaction where Cys 101 attacks Cys 98 forming a disulphide bond between Cys 101 and Cys 98. This leaves both the sulfurs of the original disulphide bond reduced.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp95A | proton donor |
| Cys101A | nucleophile |
| Cys98A | electrophile |
| Cys101A | proton donor |
| Cys98A | electrofuge |


 Download:
Download:  Download:
Download: