H+-transporting two-sector ATPase (F-type, bacterial)
The F0 subunit of E.coli ATP synthase is involved in proton translocation across the membrane from the periplasm to the cytoplasm. Structurally it shows a high degree of similarity to the mitochondrial and cytoplasmic versions of the enzyme, indicating that a common mechanism of ATP synthesis is used by all. The ATP synthase can also function in the reverse direction, where ATP hydrolysis by the F1 (soluble) subunit is coupled to movement of protons across the membrane against the concentration gradient rather than with it. The components of the ATP synthase F0 subunit are one a, 2 b and 12 c units which interact together to allow rotation to occur, driving ATP synthesis.
Reference Protein and Structure
- Sequences
-
P68699

P0AB98
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1c17
- A1C12 SUBCOMPLEX OF F1FO ATP SYNTHASE
(solution nmr
Å)



- Catalytic CATH Domains
-
1.20.20.10
 1.20.120.220
1.20.120.220  (see all for 1c17)
(see all for 1c17)
Enzyme Reaction (EC:7.1.2.2)
Enzyme Mechanism
Introduction
The synthesis of ATP occurs through proton translocation across the membrane. A proton is picked up by Asn 214 on the periplasmic side where the H+ concentration is high. This is then passed to Asp 61, causing the breaking of a salt bridge between this residue and Arg 210. As a result, the Asp 61 residue moves away from the Arg 210, causing the helix to move with it and the rotation of around 30 degrees relative to the a domain which leads to conformational changes in the F1 subunit and consequential ATP synthesis. The proton is passed from Asp 61 to the cytoplasm via Ser 206, so that the Asp 61 can accept another proton from the periplasm and continue the cycle.
Catalytic Residues Roles
| UniProt | PDB* (1c17) | ||
| Asn214 | Asn214(120)M | Picks up proton from periplasmic side of the ATP synthase and passes it to Asp 61 to cause conformational change. | proton acceptor |
| Ser206 | Ser206(112)M | Picks up proton from Asp 61 and transfers it to the cytoplasmic side of the ATP synthase, thus allowing Asp 61 to return to its original position. | proton relay, proton acceptor, proton donor |
| Asp61 | Asp61L | Accepts proton causing the change in the salt bridge with Arg 210 that results in rotation of the c subcomplex and the consequential conformational change that leads to ATP synthesis. | proton acceptor, electrostatic stabiliser, proton donor |
| Arg210 | Arg210(116)M | By forming a salt bridge to the deprotonated form of Asp 61, modifies the pKa such that protonation results in the breaking of this salt bridge and the rotation of the c subcomplex relative to the a subcomplex. | electrostatic stabiliser |
Chemical Components
proton transfer, overall reactant used, native state of enzyme regenerated, overall product formedReferences
- Dmitriev OY et al. (2002), Biochemistry, 41, 5537-5547. Structure of Ala24/Asp61 → Asp24/Asn61 Substituted SubunitcofEscherichia coliATP Synthase: Implications for the Mechanism of Proton Transport and Rotary Movement in the FoComplex†. DOI:10.1021/bi012198l. PMID:11969414.
- Mashkovtseva E et al. (2013), Math Biosci, 243, 117-125. Combined mathematical methods in the description of the F(o)F(1)-ATP synthase catalytic cycle. DOI:10.1016/j.mbs.2013.02.013. PMID:23499574.
- Mukherjee S et al. (2012), Proc Natl Acad Sci U S A, 109, 14876-14881. Realistic simulations of the coupling between the protomotive force and the mechanical rotation of the F0-ATPase. DOI:10.1073/pnas.1212841109. PMID:22927379.
- Aksimentiev A et al. (2004), Biophys J, 86, 1332-1344. Insights into the Molecular Mechanism of Rotation in the Fo Sector of ATP Synthase. DOI:10.1016/s0006-3495(04)74205-8. PMID:14990464.
- Rastogi VK et al. (1999), Nature, 402, 263-268. Structural changes linked to proton translocation by subunit c of the ATP synthase. DOI:10.1038/46224. PMID:10580496.
- Valiyaveetil FI et al. (1997), J Biol Chem, 272, 32635-32641. On the Role of Arg-210 and Glu-219 of Subunit a in Proton Translocation by the Escherichia coliF0F1-ATP Synthase. DOI:10.1074/jbc.272.51.32635. PMID:9405480.
- Kluge C et al. (1993), Biochemistry, 32, 10378-10386. Kinetics of inactivation of the F1F0 ATPase of Propionigenium modestum by dicyclohexylcarbodiimide in relationship to hydrogen ion and sodium concentration: Probing the binding site for the coupling ions. DOI:10.1021/bi00090a013. PMID:8399181.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg210(116)M | electrostatic stabiliser |
| Asp61L | electrostatic stabiliser |
| Asn214(120)M | proton acceptor |
Chemical Components
proton transfer, overall reactant used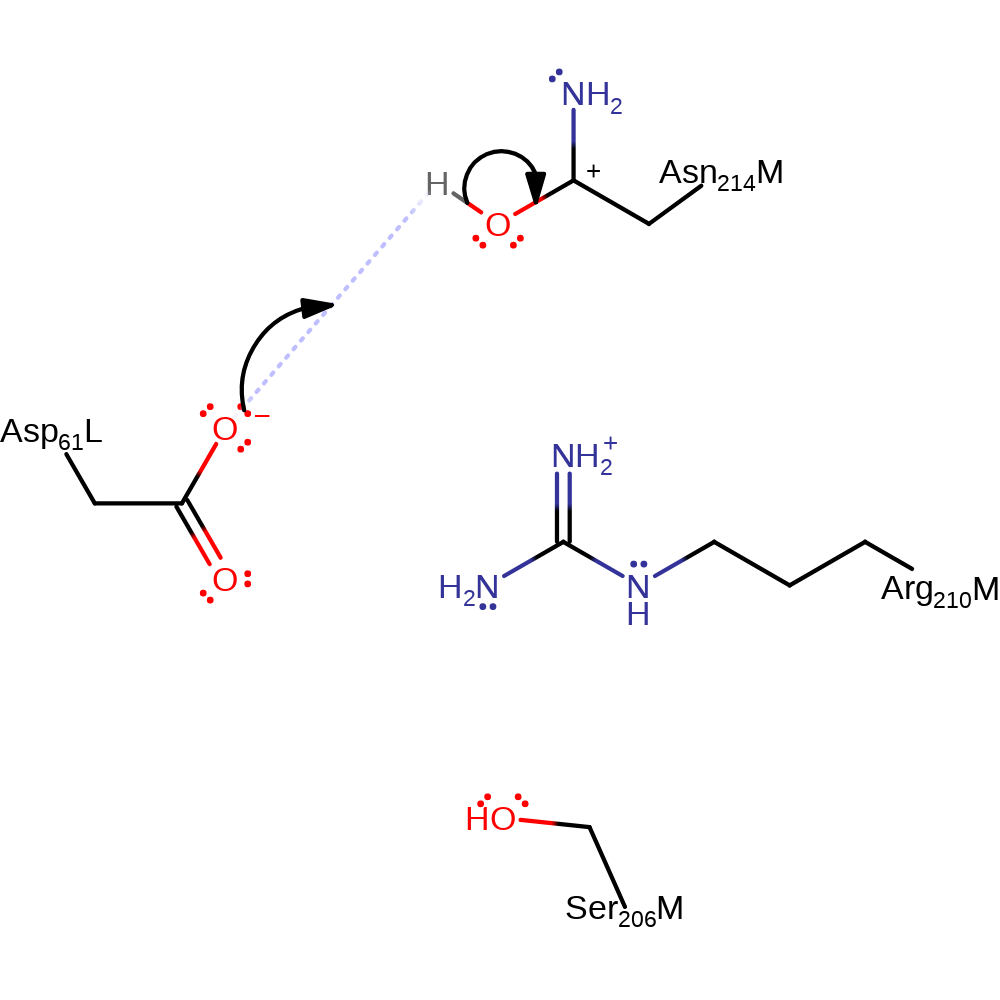
Step 2. Asp 61 (L) accepts a proton from Asn 214 (M) which results in the breaking of a salt bridge between this residue and Arg 210. As a result, the Asp 61 residue moves away from the Arg 210, causing the helix to move with it and the rotation of around 30 degrees relative to the a domain.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg210(116)M | electrostatic stabiliser |
| Asp61L | proton acceptor |
Chemical Components
proton transfer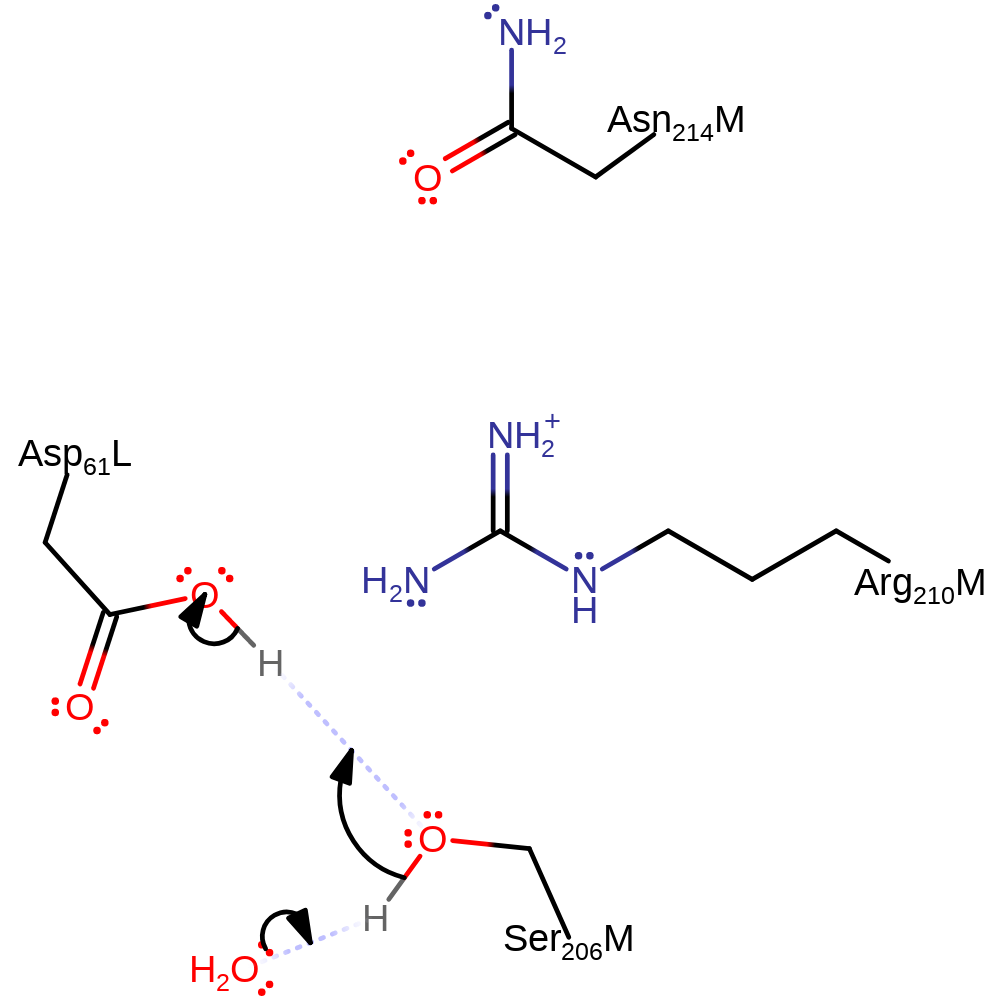
Step 3. The proton is transferred from Asp 61 (L) to Ser 206 (M) which then releases a proton into the cytoplasm. As result the helix containing Asp 61 (M) rotates back to its original conformation so that it is ready to receive another proton.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg210(116)M | electrostatic stabiliser |
| Asp61L | proton donor |
| Ser206(112)M | proton donor, proton acceptor, proton relay |






 Download:
Download: