Galactarate dehydratase (D-threo-forming)
Galactarate dehydratase (D-threo-forming) enzymes catalyse the dehydration of galactarate. The product of this reaction is the enantiomer of the product of the galactarate dehydratase reaction catalysed by the L-talarate/galactarate dehydratase family in the mandelate racemase subgroup.
Reference Protein and Structure
- Sequence
-
Q8EMJ9
 (4.2.1.158)
(4.2.1.158)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Oceanobacillus iheyensis HTE831 (Bacteria)

- PDB
-
3es7
- Crystal structure of divergent enolase from Oceanobacillus Iheyensis complexed with Mg and L-malate.
(1.9 Å)



- Catalytic CATH Domains
-
3.30.390.10
 3.20.20.120
3.20.20.120  (see all for 3es7)
(see all for 3es7)
- Cofactors
- Magnesium(2+) (2)
Enzyme Mechanism
Introduction
A Tyr-Arg dyad is responsible for the abstraction of the proton alpha to the carbonyl group in the first step of a classical enolase reaction. A second tyrosine acts as the general acid in the second step, which results in the elimination of a water molecule. The final step of the reaction is an assisted keto-enol tautomerisation that is thought to be initiated by Tyr90, and regenerates the enzyme's ground state.
Catalytic Residues Roles
| UniProt | PDB* (3es7) | ||
| Arg162 | Arg162A | Part of the Arg-Tyr dyad, it s responsible for perturbing the pKa of Tyr164 such that it can act as the initial general base. | modifies pKa, electrostatic stabiliser |
| Tyr164 | Tyr164A | Acts as a general acid/base. This tyrosine is activated as part of an Arg-Tyr dyad. It is responsible for the initial deprotonation if the galactarate C2 atom. | proton acceptor, proton donor |
| His246, Asp193, Glu221 | His246A, Asp193A, Glu221A | Forms the magnesium 2 binding site. This is the classical enolase metal binding site. | metal ligand |
| Thr297, Asp42, His45 | Thr297A, Asp42A, His45A | Form the magnesium 1 binding site. | metal ligand |
| Tyr90 | Tyr90A | Acts as a general acid/base. This tyrosine is on the opposite side of the substrate to Tyr164 and is responsible for facilitating the departure of beta-hydroxide leaving group. It also acts as the acid catalyst for ketonization of enol intermediate. | proton acceptor, proton donor |
Chemical Components
assisted keto-enol tautomerisation, dehydration, unimolecular elimination by the conjugate base, native state of enzyme regeneratedReferences
- Rakus JF et al. (2009), Biochemistry, 48, 11546-11558. Computation-Facilitated Assignment of the Function in the Enolase Superfamily: A Regiochemically Distinct Galactarate Dehydratase fromOceanobacillus iheyensis,. DOI:10.1021/bi901731c. PMID:19883118.
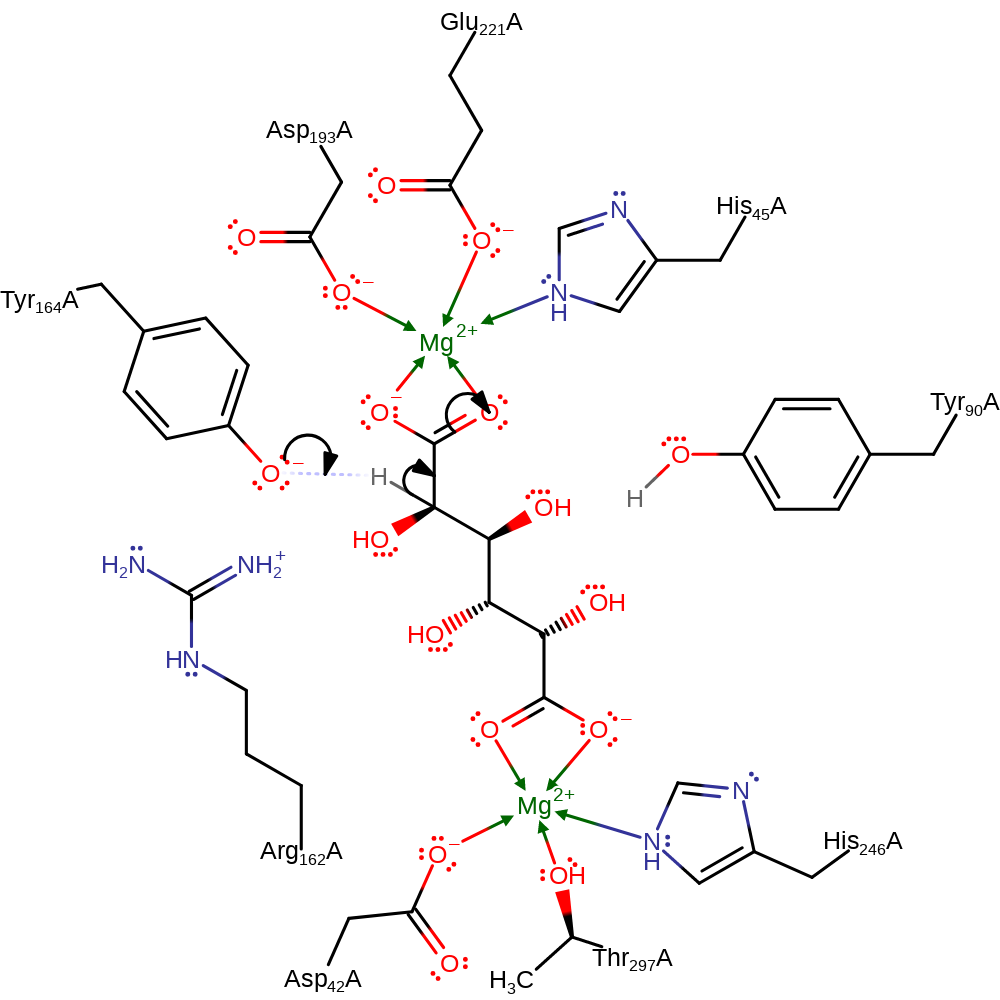
Step 1. Tyr164, activated by Arg162, abstracts the C2 proton from the galactarate substrate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg162A | modifies pKa |
| Asp193A | metal ligand |
| Glu221A | metal ligand |
| His45A | metal ligand |
| Asp42A | metal ligand |
| Thr297A | metal ligand |
| His246A | metal ligand |
| Arg162A | electrostatic stabiliser |
| Tyr164A | proton acceptor |
Chemical Components
assisted keto-enol tautomerisation
Step 2. The negatively charged intermediate collapses, eliminating water (with concomitant deprotonation of Tyr90).
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp193A | metal ligand |
| Glu221A | metal ligand |
| His45A | metal ligand |
| Asp42A | metal ligand |
| Thr297A | metal ligand |
| His246A | metal ligand |
| Tyr90A | proton donor |
Chemical Components
assisted keto-enol tautomerisation, dehydration, ingold: unimolecular elimination by the conjugate base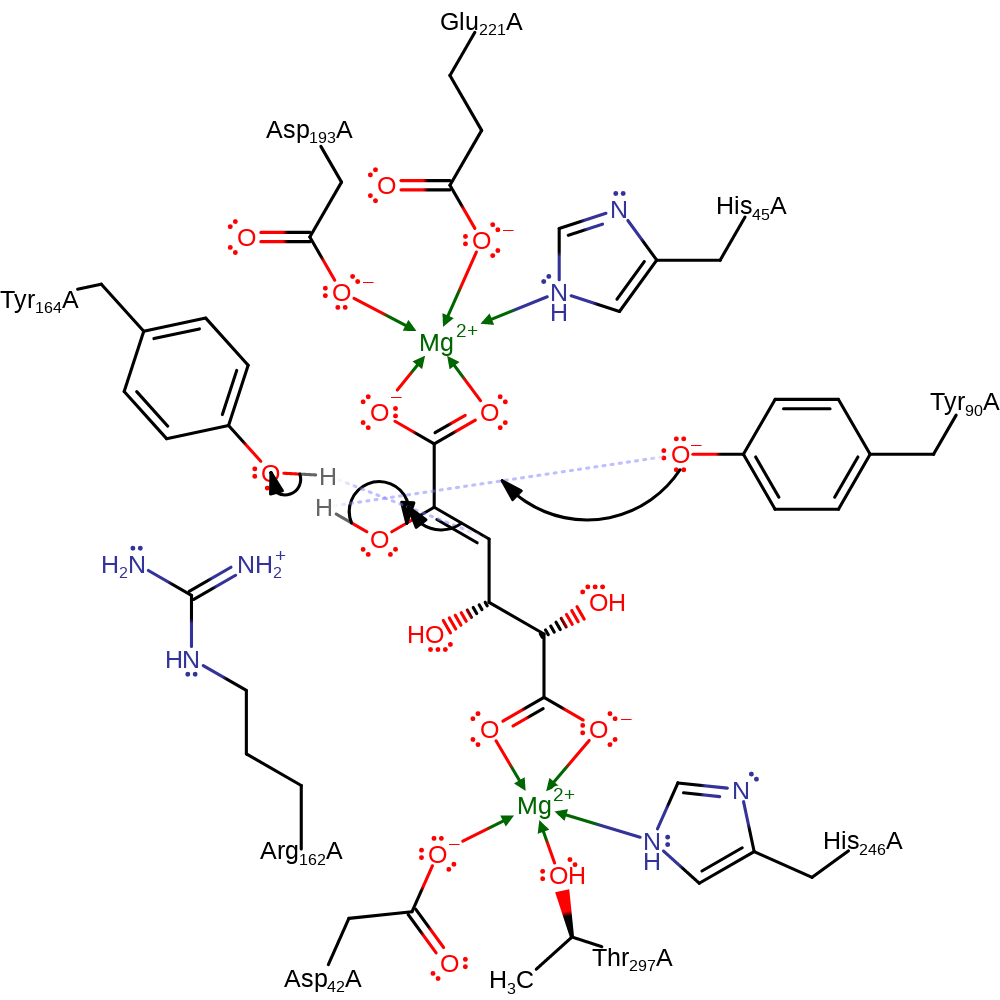
Step 3. Tyr90 deprotonates the OH group of the interemdiate in an assisted keto-enol tautomerisation. This results in the regeneration of the active site and production of the final product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg162A | modifies pKa |
| Asp193A | metal ligand |
| Glu221A | metal ligand |
| His45A | metal ligand |
| Asp42A | metal ligand |
| Thr297A | metal ligand |
| His246A | metal ligand |
| Tyr164A | proton donor |
| Tyr90A | proton acceptor |



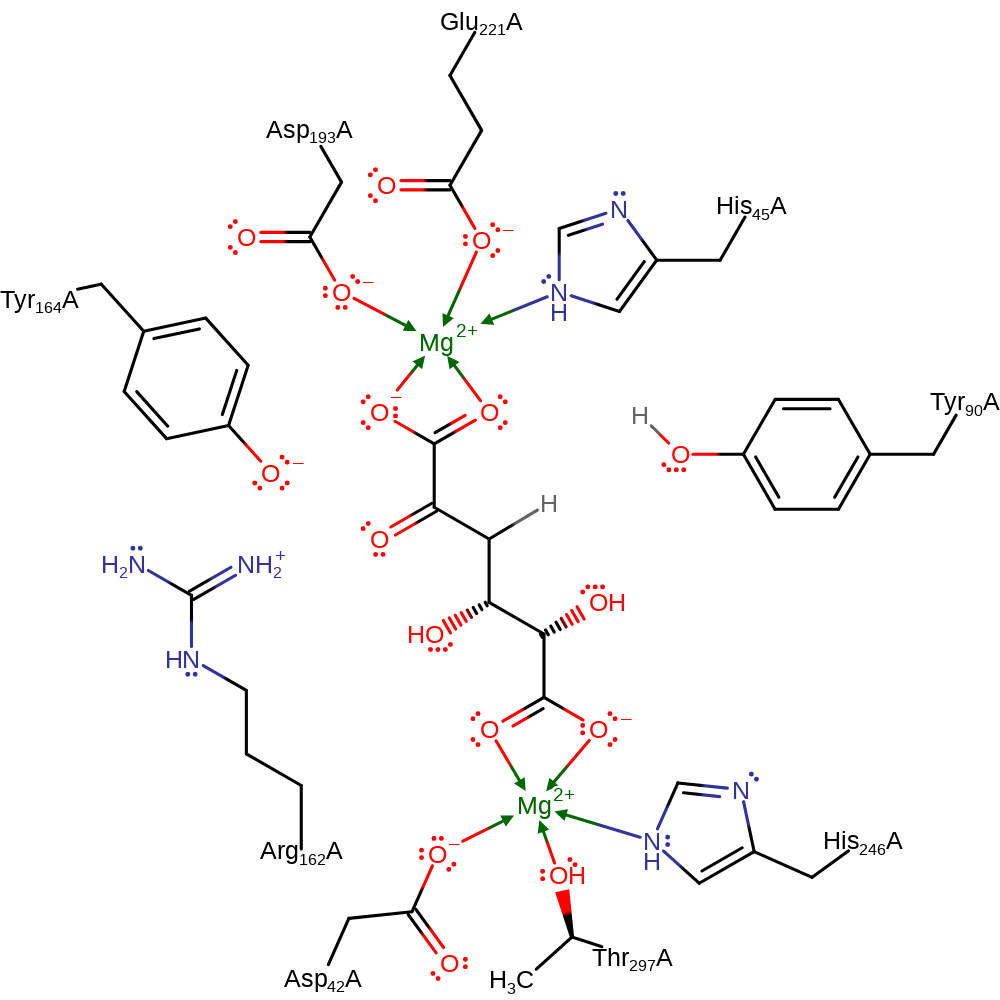 Download:
Download: