Serine/threonine-protein phosphatase 5
Serine/threonine protein phosphatase 5 (PP5) from Homo sapiens hydrolyses a phosphoprotein substrate into a protein and a phosphate group. It is implicated in wide ranging cellular processes, including apoptosis, differentiation, DNA damage response, cell survival, regulation of ion channels or circadian rhythms, in response to steroid and thyroid hormones, calcium, fatty acids, TGF-beta as well as oxidative and genotoxic stresses. It is a member of the PPP gene family of protein phosphatases that is widely expressed in mammalian tissues.
Reference Protein and Structure
- Sequence
-
P53041
 (3.1.3.16)
(3.1.3.16)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Homo sapiens (Human)

- PDB
-
1s95
- Structure of serine/threonine protein phosphatase 5
(1.6 Å)



- Catalytic CATH Domains
-
3.60.21.10
 (see all for 1s95)
(see all for 1s95)
- Cofactors
- Manganese(2+) (2)
Enzyme Reaction (EC:3.1.3.16)
Enzyme Mechanism
Introduction
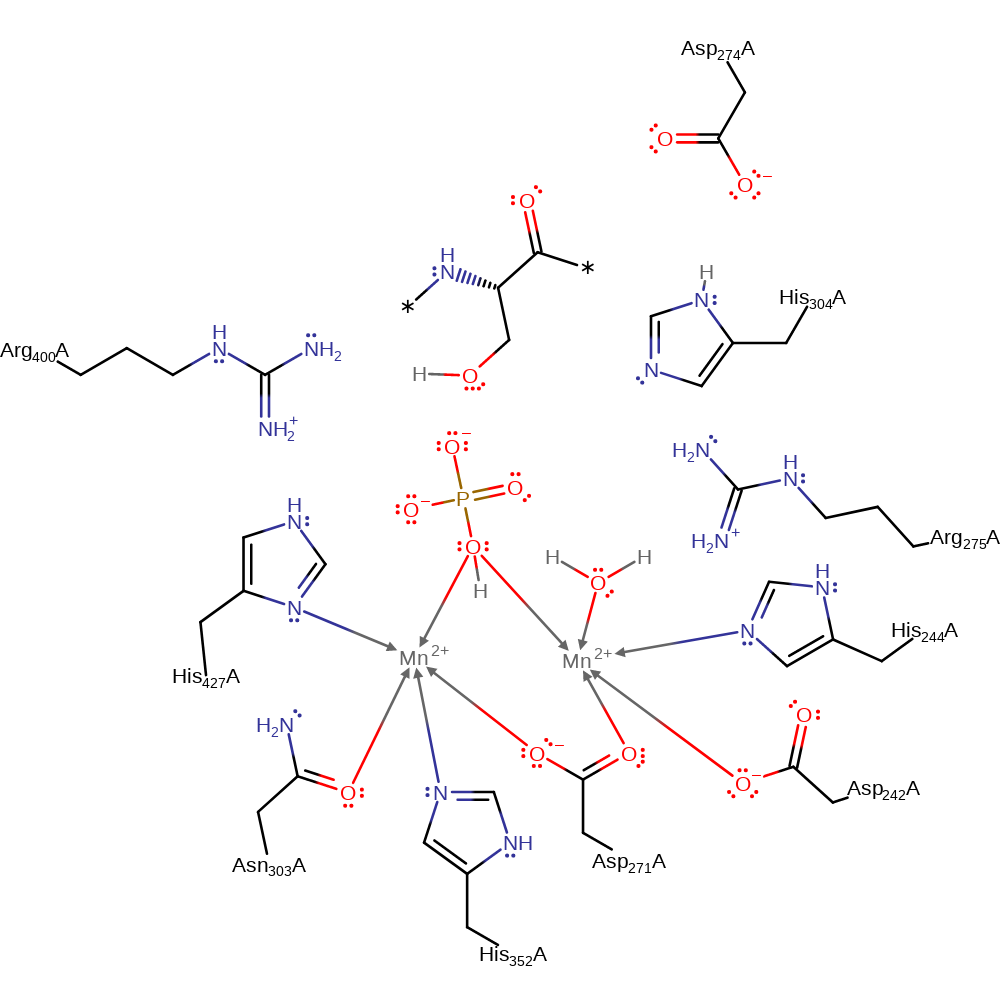
DFT calculations suggest that the reaction proceeds via an in-line concerted transition state, without intermediates. A water molecule is activated by two Mn(II) ions and His427 which lower its pKa such that it is present as a hydroxide ion. This hydroxide ion performs a nucleophilic attack on the phosphorus atom of the substrate, causing the P-O4 bond to break. The transition state is stabilised by Arg275 (H bonding to O1 and O4), Arg400 (H bonding to O3) and Asn303 (H bonding to O2). Asp274 hydrogen bonds to His304, lowering its pKa, facilitating deprotonation of His304. The leaving alcohol group is protonated by His304 and His304 is protonated by Asp274.
Catalytic Residues Roles
| UniProt | PDB* (1s95) | ||
| Asp242, His244, Asp271 | Asp242(76)A, His244(78)A, Asp271(105)A | Forms the manganese 1 binding site. | metal ligand |
| Asp274 | Asp274(108)A | Hydrogen bonds to His 304, lowering its pKa, aiding His 304 to donate a proton to the leaving group. | activator, proton donor |
| His304 | His304(138)A | Acts as an acid by donating a proton to the leaving alcohol group. | proton relay, proton acceptor, proton donor |
| Arg275 | Arg275(109)A | Hydrogen bonds to atoms O1 and O4, helping to stabilise the transition state. | transition state stabiliser |
| Asn303 | Asn303(137)A | Hydrogen bonds to atom O2 of the substrate, helping to stabilise the transition state. | metal ligand, transition state stabiliser |
| Arg400 | Arg400(234)A | Hydrogen bonds to atom O3 of the substrate, helping to stabilise the transition state. | transition state stabiliser |
| Asp271, His352, Asn303, His427 | Asp271(105)A, His352(186)A, Asn303(137)A, His427(261)A | Forms the manganese 2 binding site. | metal ligand |
| His427 | His427(261)A | Helps lower the pKa of a bound water molecule to such an extent as to cause it to 'spontaneously deprotonate'. | activator, metal ligand |
Chemical Components
bimolecular nucleophilic substitution, proton transfer, overall product formedReferences
- Ribeiro AJ et al. (2013), Chemistry, 19, 14081-14089. The Catalytic Mechanism of Protein Phosphatase 5 Established by DFT Calculations. DOI:10.1002/chem.201301565. PMID:24014428.
- McConnell JL et al. (2009), Mol Pharmacol, 75, 1249-1261. Targeting Protein Serine/Threonine Phosphatases for Drug Development. DOI:10.1124/mol.108.053140. PMID:19299564.
- Shi Y (2009), Cell, 139, 468-484. Serine/Threonine Phosphatases: Mechanism through Structure. DOI:10.1016/j.cell.2009.10.006. PMID:19879837.
- Hinds TD Jr et al. (2008), Int J Biochem Cell Biol, 40, 2358-2362. Protein phosphatase 5. DOI:10.1016/j.biocel.2007.08.010. PMID:17951098.
- Swingle MR et al. (2004), J Biol Chem, 279, 33992-33999. Structural Basis for the Catalytic Activity of Human Serine/Threonine Protein Phosphatase-5. DOI:10.1074/jbc.m402855200. PMID:15155720.

Step 1. The bound hydroxide performs a nucleophilic attack on the phosphate group causing it to be cleaved from the main substrate molecule. The oxygen of the dephosphorylated protein deprotonates His304 which in turn deprotonates Asp274.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp242(76)A | metal ligand |
| His244(78)A | metal ligand |
| Asp271(105)A | metal ligand |
| Asn303(137)A | metal ligand |
| His352(186)A | metal ligand |
| His427(261)A | metal ligand |
| Asp274(108)A | activator |
| Arg400(234)A | transition state stabiliser |
| His427(261)A | activator |
| Asn303(137)A | transition state stabiliser |
| Arg275(109)A | transition state stabiliser |
| His304(138)A | proton donor, proton acceptor |
| Asp274(108)A | proton donor |
| His304(138)A | proton relay |




 Download:
Download: