Asparaginase
Asparaginase (EC 3.5.1.1) and glutaminaase (EC 3.5.1.38) remove amino groups from asparagine and glutamine respectively leaving aspartate and glutamate. This entry covers both enzymes, though is derived from asparaginase. Enzymes from both EC families have some activity in the other. Two forms exist in E.coli, type I is cytoplasmic, type II is periplasmic. The type II form has a higher affinity for the substrate and its production is induced under anaerobic conditions when amino acids are the primary carbon source. Type II L-asparaginase is the most studied, because it is an important anti-cancer drug due to its ability to lower the levels of asparagine available to cancerous cells lacking in asparaginase synthase.
Reference Protein and Structure
- Sequence
-
P00805
 (3.5.1.1)
(3.5.1.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
3eca
- CRYSTAL STRUCTURE OF ESCHERICHIA COLI L-ASPARAGINASE, AN ENZYME USED IN CANCER THERAPY
(2.4 Å)



- Catalytic CATH Domains
-
3.40.50.40
 3.40.50.1170
3.40.50.1170  (see all for 3eca)
(see all for 3eca)
Enzyme Reaction (EC:3.5.1.1)
Enzyme Mechanism
Introduction
In the double displacement (ping-pong) mechanism proposal, the reaction proceeds via a nucleophilic attack of Thr12 on the CG/CD of asparagine/glutamine forming a covalent intermediate. After the intermediate is formed, the amine group is eliminated, followed by a second nucleophilic attack on the substrate, this time by a water molecule, which ultimately leads to the collapse of the covalent intermediate and regeneration of the active site. The existence of a crystallographic structure (PDB ID:6v26) of an acyl-enzyme intermediate (post ammonia elimination) is a strong piece of experimental evidence in favour of this mechanism. Although computational studies did not support a previous version of this mechanism, it has been suggested that two water molecules present in the active site but not taken into account in the computational studies might account for additional stabilisation.
Catalytic Residues Roles
| UniProt | PDB* (3eca) | ||
| Thr34 | Thr12A | Acts as a catalytic nucleophile. | covalently attached, nucleofuge, nucleophile, proton acceptor, proton donor |
| Thr111 (main-N) | Thr89A (main-N) | The main chain amide forms part of the oxyanion hole along with the active site water molecule. | electrostatic stabiliser |
| Tyr47 | Tyr25A | Activates Thr12 via proton abstraction. | proton relay, increase nucleophilicity, proton acceptor, proton donor |
| Thr111, Asp112, Lys184 | Thr89A, Asp90A, Lys162A | Acts as the general acid/base. | proton relay, proton acceptor, proton donor |
Chemical Components
bimolecular nucleophilic addition, proton transfer, unimolecular elimination by the conjugate baseReferences
- Lubkowski J et al. (2020), Biochemistry, 59, 1927-1945. Mechanism of Catalysis by l-Asparaginase. DOI:10.1021/acs.biochem.0c00116. PMID:32364696.
- Borek D et al. (2014), FEBS J, 281, 4097-4111. Crystal structure of active site mutant of antileukemicl-asparaginase reveals conserved zinc-binding site. DOI:10.1111/febs.12906. PMID:25040257.
- Gesto DS et al. (2013), J Am Chem Soc, 135, 7146-7158. Unraveling the Enigmatic Mechanism ofl-Asparaginase II with QM/QM Calculations. DOI:10.1021/ja310165u. PMID:23544711.
- Aung HP et al. (2000), Biochim Biophys Acta, 1481, 349-359. Dynamics of a mobile loop at the active site of Escherichia coli asparaginase. DOI:10.1016/s0167-4838(00)00179-5. PMID:11018727.
- Lubkowski J et al. (1996), Eur J Biochem, 241, 201-207. Crystal Structure and Amino Acid Sequence of Wolinella Succinogenesl-Asparaginase. DOI:10.1111/j.1432-1033.1996.0201t.x. PMID:8898907.
- Palm GJ et al. (1996), FEBS Lett, 390, 211-216. A covalently bound catalytic intermediate inEscherichia coliasparaginase : Crystal structure of a Thr-89-Val mutant. DOI:10.1016/0014-5793(96)00660-6. PMID:8706862.
- Swain AL et al. (1993), Proc Natl Acad Sci U S A, 90, 1474-1478. Crystal structure of Escherichia coli L-asparaginase, an enzyme used in cancer therapy. DOI:10.1073/pnas.90.4.1474. PMID:8434007.
- Harms E et al. (1991), FEBS Lett, 285, 55-58. A catalytic role for threonine-12 of E. coli asparaginase II as established by site-directed mutagenesis. PMID:1906013.
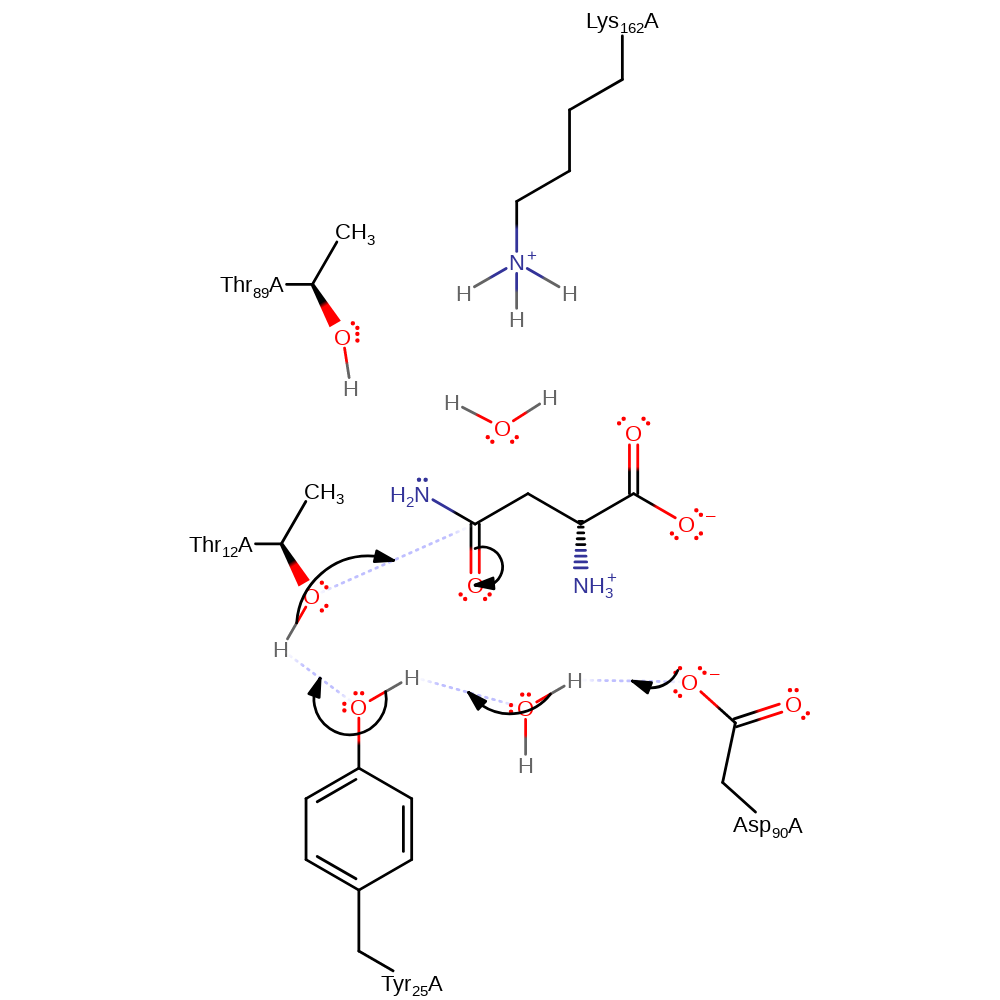
Step 1. Thr12 is activated by Tyr25 and Asp90 and performs a nucleophilic attack on the substrate, forming the covalent intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp90A | proton acceptor |
| Tyr25A | proton acceptor, proton donor, proton relay |
| Thr12A | nucleophile, proton donor |
| Asp90A | increase nucleophilicity |
| Tyr25A | increase nucleophilicity |
| Thr89A (main-N) | electrostatic stabiliser |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer
Step 2. Ammonia is released while accepting a proton from Thr89 which in turn deprotonates Lys162.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys162A | proton acceptor |
| Thr12A | covalently attached |
| Thr89A | proton acceptor, proton donor, proton relay |
| Thr89A (main-N) | electrostatic stabiliser |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base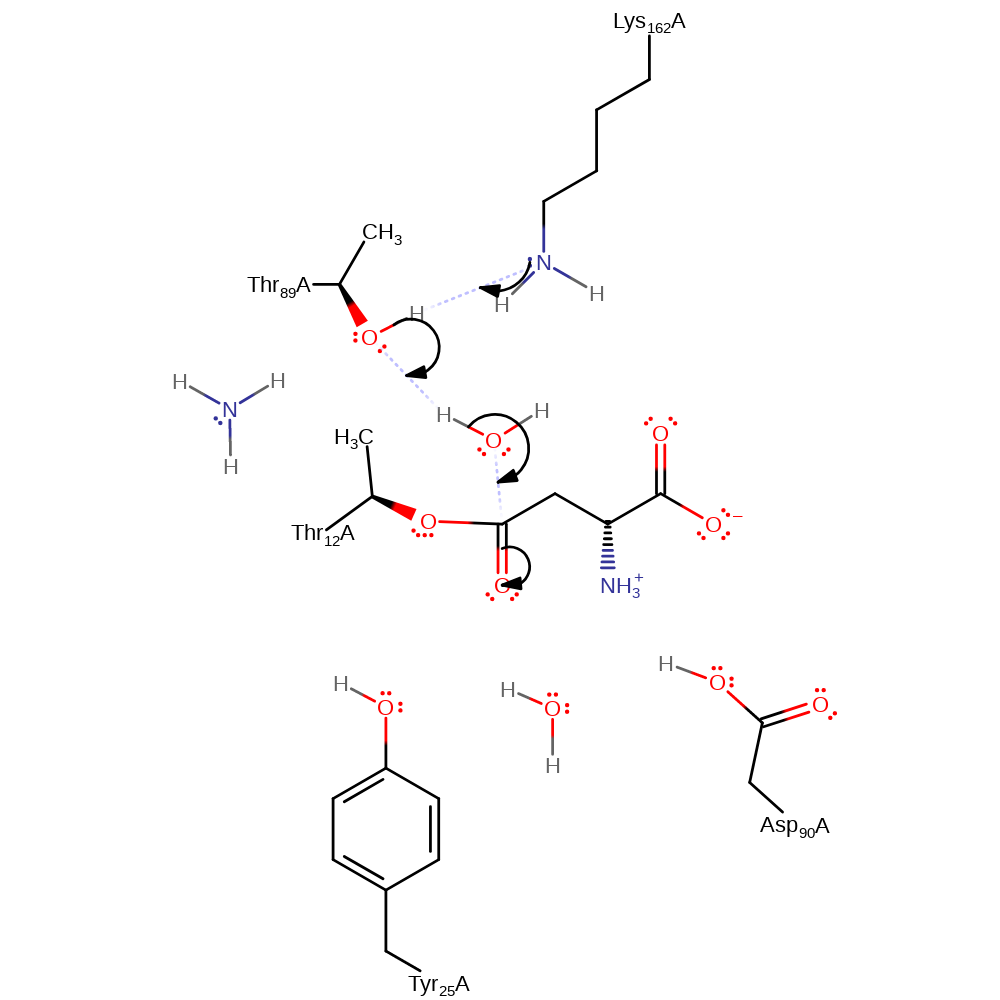
Step 3. A water molecule performs a nucleophilic attack on the acyl-enzyme intermediate, forming again a tetrahedral intermediate. A proton from the nucleophilic water molecule is transferred to Thr89 which in turn looses a proton to Lys162.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys162A | proton acceptor |
| Thr89A | proton acceptor, proton donor, proton relay |
| Thr89A (main-N) | electrostatic stabiliser |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer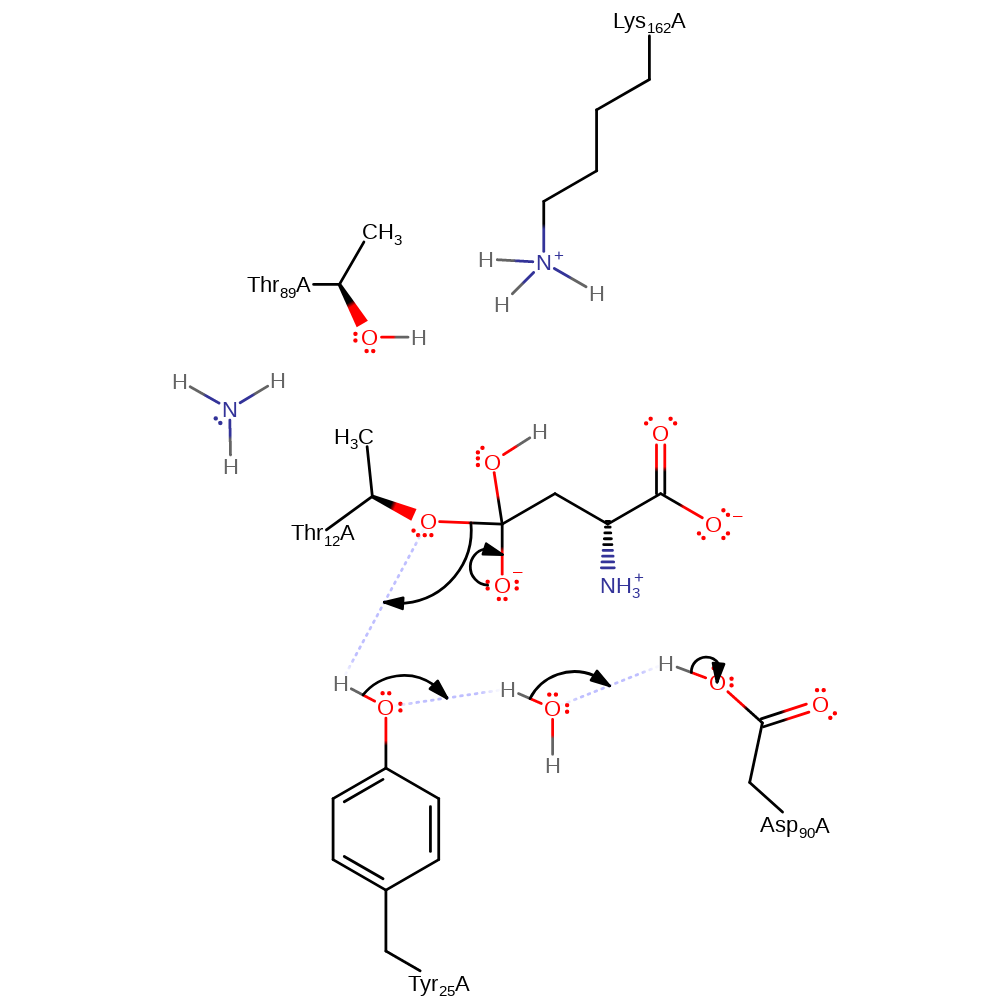
Step 4. The covalent intermediate collapses, as Thr12 is protonated by Tyr25, which in turn is re-protonated by Asp90 through a bridging water. The final product is formed.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr12A | nucleofuge, proton acceptor |
| Tyr25A | proton acceptor, proton donor, proton relay |
| Asp90A | proton donor |
| Thr89A (main-N) | electrostatic stabiliser |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate baseIntroduction
In this direct displacement mechanism proposal, the Thr89-Lys162-Asp90 triad activates a water molecule to perform the nucleophilic attack in the first step of reaction, while the second triad, Thr12-Tyr25-Glu283, stabilizes the tetrahedral intermediate formed in the first step. The Thr89-Lys162-Asp90 triad is situated in a rigid part of the structure and mutational and crystallographic studies indicate that all three residues, which are absolutely conserved, are involved in the catalytic reaction.
Catalytic Residues Roles
| UniProt | PDB* (3eca) | ||
| Thr34, Glu305, Tyr47 | Thr12A, Glu283C, Tyr25A | Forms the Thr-Tyr-Glu triad that is responsible for stabilising the intermediates of the reaction. | electrostatic stabiliser |
| Thr111, Asp112 | Thr89A, Asp90A | Activates Lys162 for the initial proton abstraction from the water molecule. | proton acceptor, proton donor, proton relay, electrostatic stabiliser, increase basicity |
| Asp112, Lys184 | Asp90A, Lys162A | Forms the Thr-Lys-Asp triad that activates the Thr as a general acid/base. | increase basicity, electrostatic stabiliser, increase acidity |
| Lys184 | Lys162A | Acts as a general acid/base. | proton acceptor, proton donor |
Chemical Components
bimolecular nucleophilic addition, proton transfer, unimolecular elimination by the conjugate base, native state of enzyme regeneratedReferences
- Gesto DS et al. (2013), J Am Chem Soc, 135, 7146-7158. Unraveling the Enigmatic Mechanism ofl-Asparaginase II with QM/QM Calculations. DOI:10.1021/ja310165u. PMID:23544711.
- Schalk AM et al. (2016), J Biol Chem, 291, 5088-5100. Experimental Data in Support of a Direct Displacement Mechanism for Type I/II l-Asparaginases. DOI:10.1074/jbc.m115.699884. PMID:26733195.
- Borek D et al. (2014), FEBS J, 281, 4097-4111. Crystal structure of active site mutant of antileukemicl-asparaginase reveals conserved zinc-binding site. DOI:10.1111/febs.12906. PMID:25040257.
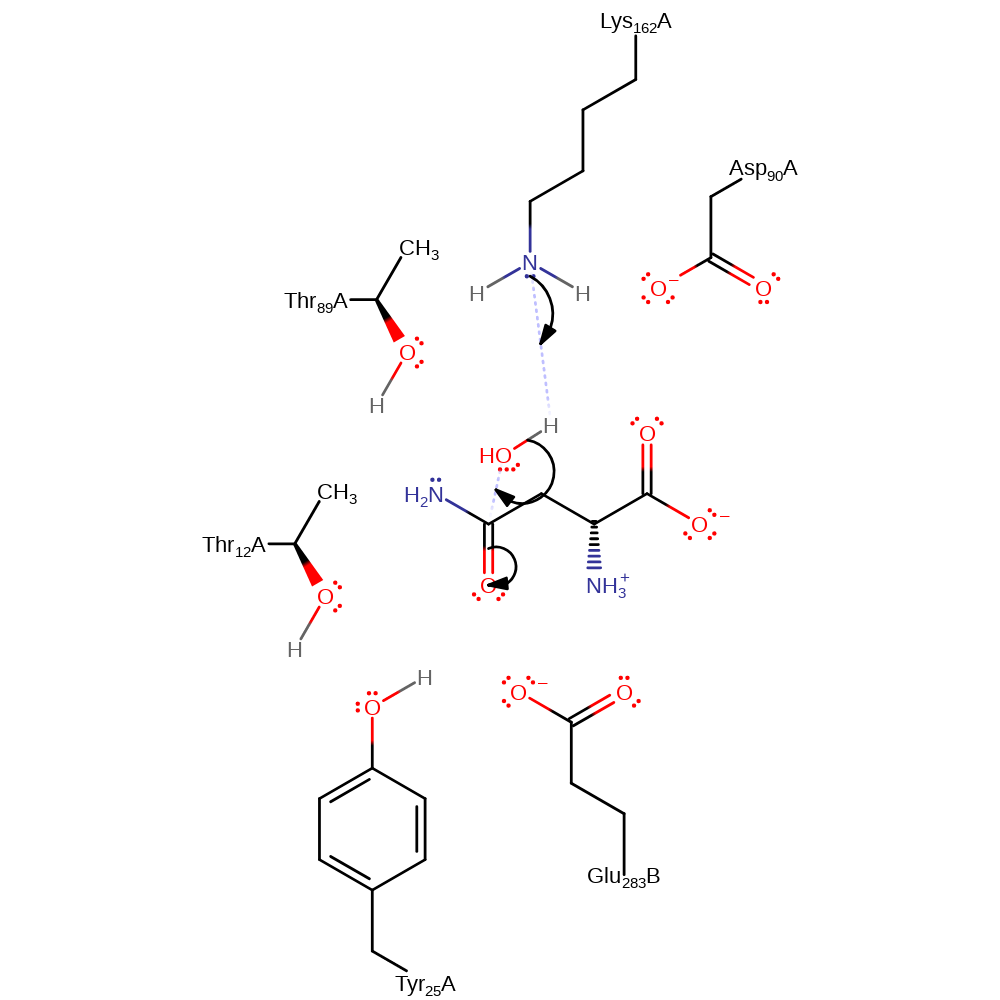
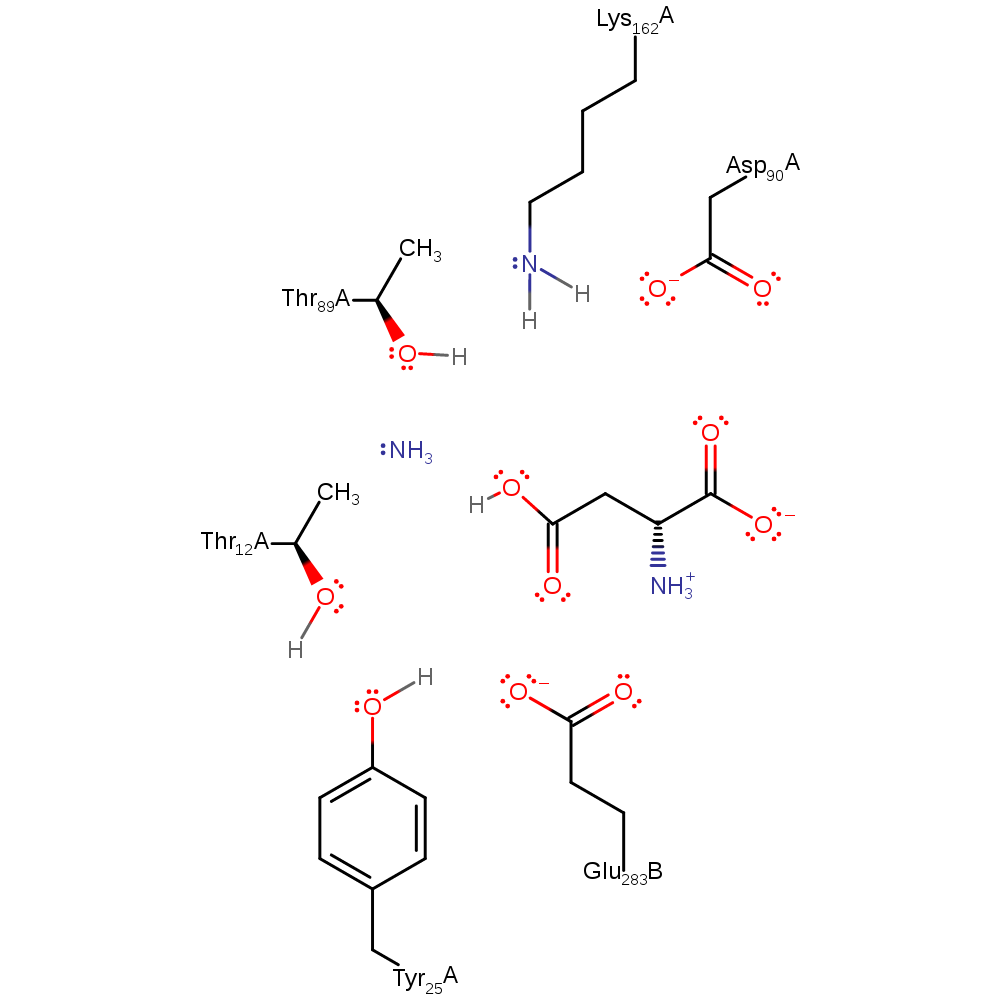
Step 1. Lys162, activated by Asp90 and Thr89, abstracts a proton from water, which initiates a nucleophilic attack on the carbonyl carbon, resulting in a negatively charged intermediate, stabilises by Thr12.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu283C | electrostatic stabiliser |
| Tyr25A | electrostatic stabiliser |
| Thr12A | electrostatic stabiliser |
| Asp90A | electrostatic stabiliser, increase basicity |
| Thr89A | increase basicity, electrostatic stabiliser |
| Lys162A | proton acceptor |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer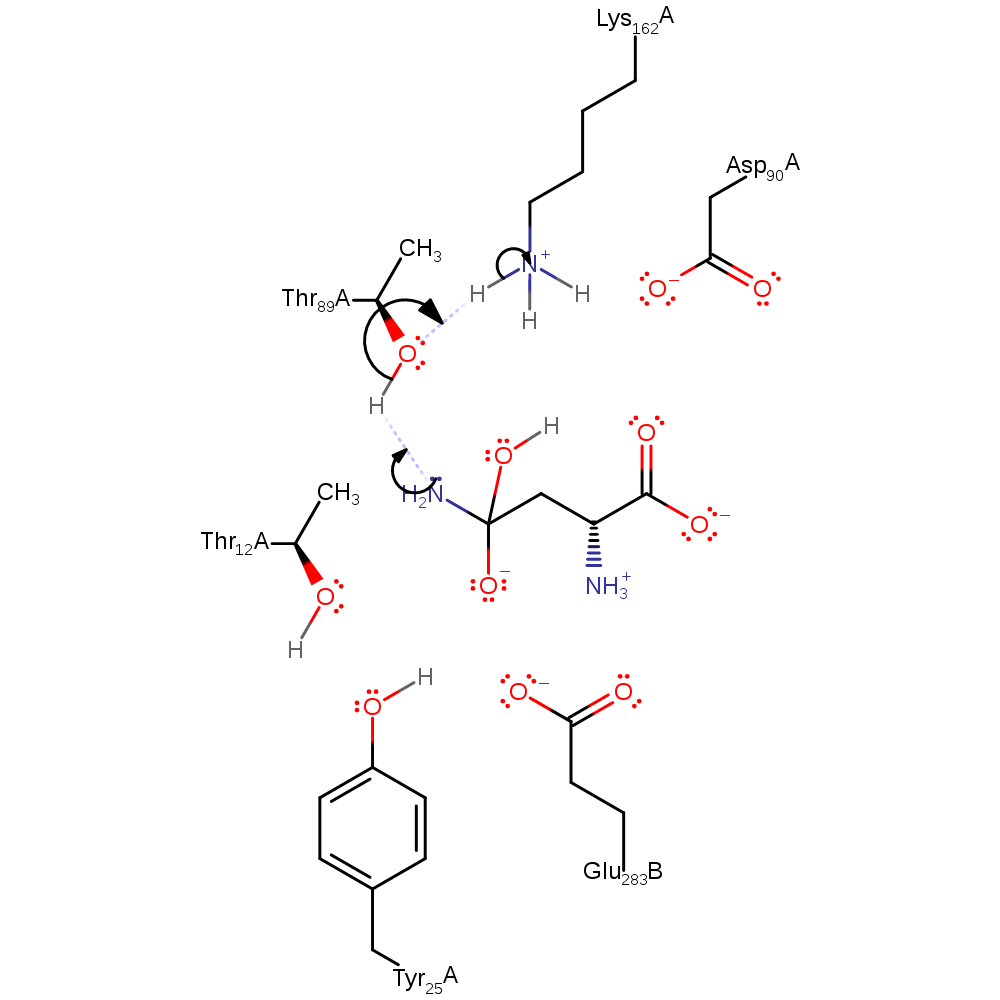
Step 2. The amine group of the intermediate abstracts a proton from Thr89, which in turn abstracts a proton from Lys162.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp90A | increase acidity |
| Thr12A | electrostatic stabiliser |
| Glu283C | electrostatic stabiliser |
| Tyr25A | electrostatic stabiliser |
| Thr89A | proton relay |
| Thr89A | proton donor, proton acceptor |
| Lys162A | proton donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transferCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr12A | electrostatic stabiliser |
| Tyr25A | electrostatic stabiliser |



 Download:
Download: 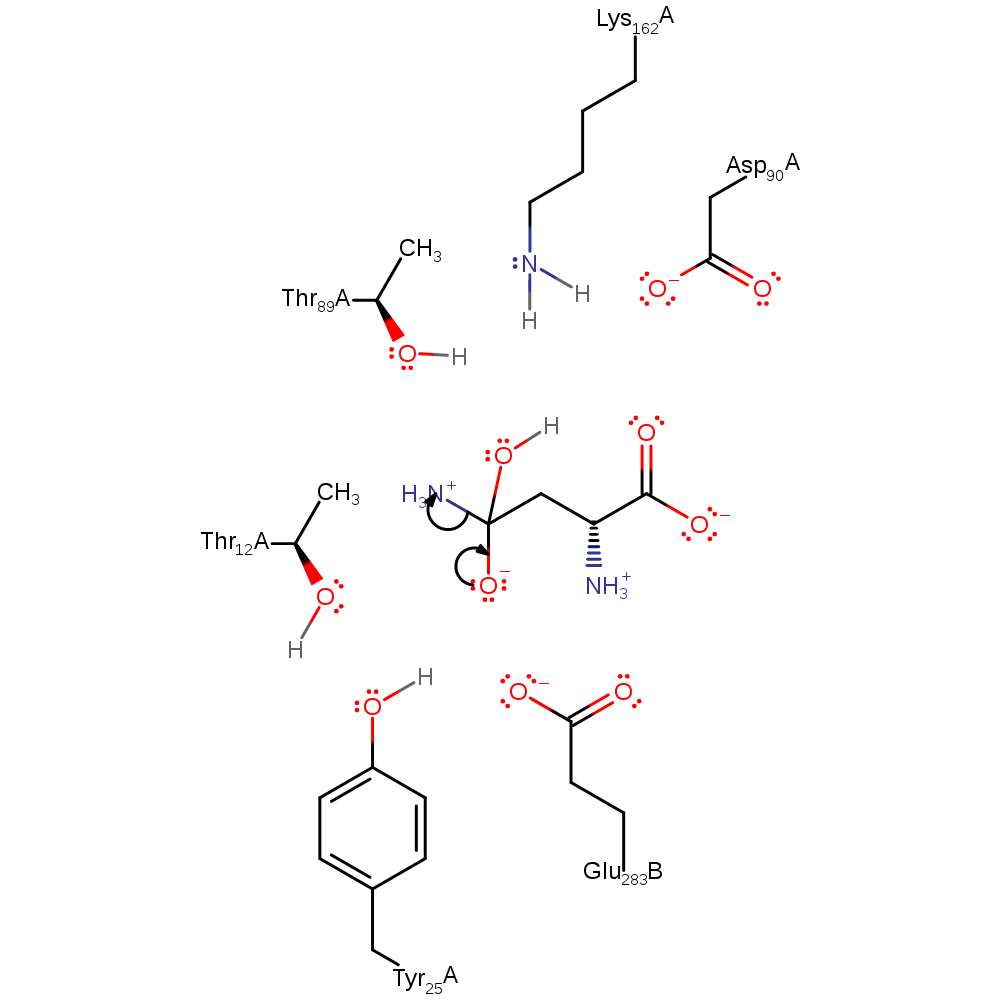
 Download:
Download: