Glucarate dehydratase
Delta-Glucarate dehyratase catalyses the dehydration of both D-glucarate and L-idarate to form 5-keto-4-deoxy-d-glucarate. Enzymes are also known to perform the glucarate to idarate and idarate to glucarate epimerization reactions
The enzyme belongs to the mechanistically diverse enolase superfamily, specifically the glucarate dehydratase subgroup (SFLD nomenclature). Enolase enzymes catalyse reactions involving the removal of an alpha proton adjacent to a carboxylate anion.
Reference Protein and Structure
- Sequence
-
P0AES2
 (4.2.1.40)
(4.2.1.40)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1ec9
- E. COLI GLUCARATE DEHYDRATASE BOUND TO XYLAROHYDROXAMATE
(2.0 Å)



- Catalytic CATH Domains
-
3.20.20.120
 (see all for 1ec9)
(see all for 1ec9)
- Cofactors
- Magnesium(2+) (1)
Enzyme Reaction (EC:4.2.1.40)
Enzyme Mechanism
Introduction
Delta-Glucarate dehyratase catalyses the dehydration of both D-Glucarate and L-Idarate to form 5-keto-4-deoxyglucarate (KDG) as well as the epimerisation of the two substrates. In the first step, the His 339 residue acts as a general base towards the C5 atom of D-Glucarate, while Lys 207 acts as a general base towards the related epimer L-Idarate. Each residue is associated with a different stereo selective function; Lys 207 acts as an S specific base, while His 339 acts as an R specific base. The enolate anion intermediate is stabilised by hydrogen bonds to residues Lys 205 and Asn 237, as well as interaction with the catalytically essential divalent Mg cation. Both the following steps, the general acid catalysed vinylogous elimination of the 4-OH group from the intermediate, generating an enol intermediate, and the general acid catalysed stereospecific tautomerisation of the enol intermediate to form the KDG product involve the His 339 residue. The use of His 339 in all three partial reactions is a good example of functional economy within enzyme catalysis.
Catalytic Residues Roles
| UniProt | PDB* (1ec9) | ||
| His339 | His339A | The residue is catalytically active in all three partial reactions involving the D-Glucarate substrate, and the second two reactions when the related epimer L-Idarate is present. The residue acts as a general base towards the H bound at the C5 of the substrate, a general acid towards the enolate intermediate, resulting the in the loss of water, and then again as a general acid towards the enol intermediate, leading to stereospecific tautomerisation and formation of the KDG product. | proton acceptor, proton donor |
| Asn237, Lys205 | Asn237A, Lys205A | The residue hydrogen bonds to, and stabilises the enolate anion intermediate. | activator, electrostatic stabiliser |
| Lys207 | Lys207A | Acts as a S stereospecific general base towards the L-Idarate substrate. | electrostatic stabiliser |
| Asp235, Glu260, Asn289 | Asp235A, Glu260A, Asn289A | Forms part of the magnesium binding site. | metal ligand |
| Asp313 | Asp313A | Activates His339 to act as the general acid/base. | modifies pKa, electrostatic stabiliser |
| Asn341 | Asn341A | Exact role unclear; could function as a general acid/base that facilitates the departure of the 4-leaving group or is essential for proper positioning of His 339. | activator |
Chemical Components
proton transfer, assisted keto-enol tautomerisation, unimolecular elimination by the conjugate base, dehydration, native state of enzyme regeneratedReferences
- Gulick AM et al. (2001), Biochemistry, 40, 10054-10062. Evolution of Enzymatic Activities in the Enolase Superfamily: Identification of the General Acid Catalyst in theActive Site ofd-Glucarate Dehydratase fromEscherichia coli†,‡. DOI:10.1021/bi010733b. PMID:11513584.
- Tian B et al. (2013), Biochemistry, 52, 5511-5513. Predicting Enzyme–Substrate Specificity with QM/MM Methods: A Case Study of the Stereospecificity ofd-Glucarate Dehydratase. DOI:10.1021/bi400546j. PMID:23901785.
- Gulick AM et al. (2000), Biochemistry, 39, 4590-4602. Evolution of Enzymatic Activities in the Enolase Superfamily: Crystallographic and Mutagenesis Studies of the Reaction Catalyzed byd-Glucarate Dehydratase fromEscherichia coli†,‡. DOI:10.1021/bi992782i. PMID:10769114.
- Hubbard BK et al. (1998), Biochemistry, 37, 14369-14375. Evolution of Enzymatic Activities in the Enolase Superfamily: Characterization of the (D)-Glucarate/Galactarate Catabolic Pathway inEscherichia coli†. DOI:10.1021/bi981124f. PMID:9772162.

Step 1. His339 abstracts a proton from the carbon alpha to the carbonyl group. ten resulting anion is stabilised through interactions with Mg(II).
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp313A | modifies pKa |
| Asn341A | activator |
| Glu260A | metal ligand |
| Asp235A | metal ligand |
| Asn289A | metal ligand |
| Asn237A | electrostatic stabiliser |
| Lys207A | electrostatic stabiliser |
| His339A | proton acceptor |
Chemical Components
proton transfer, assisted keto-enol tautomerisation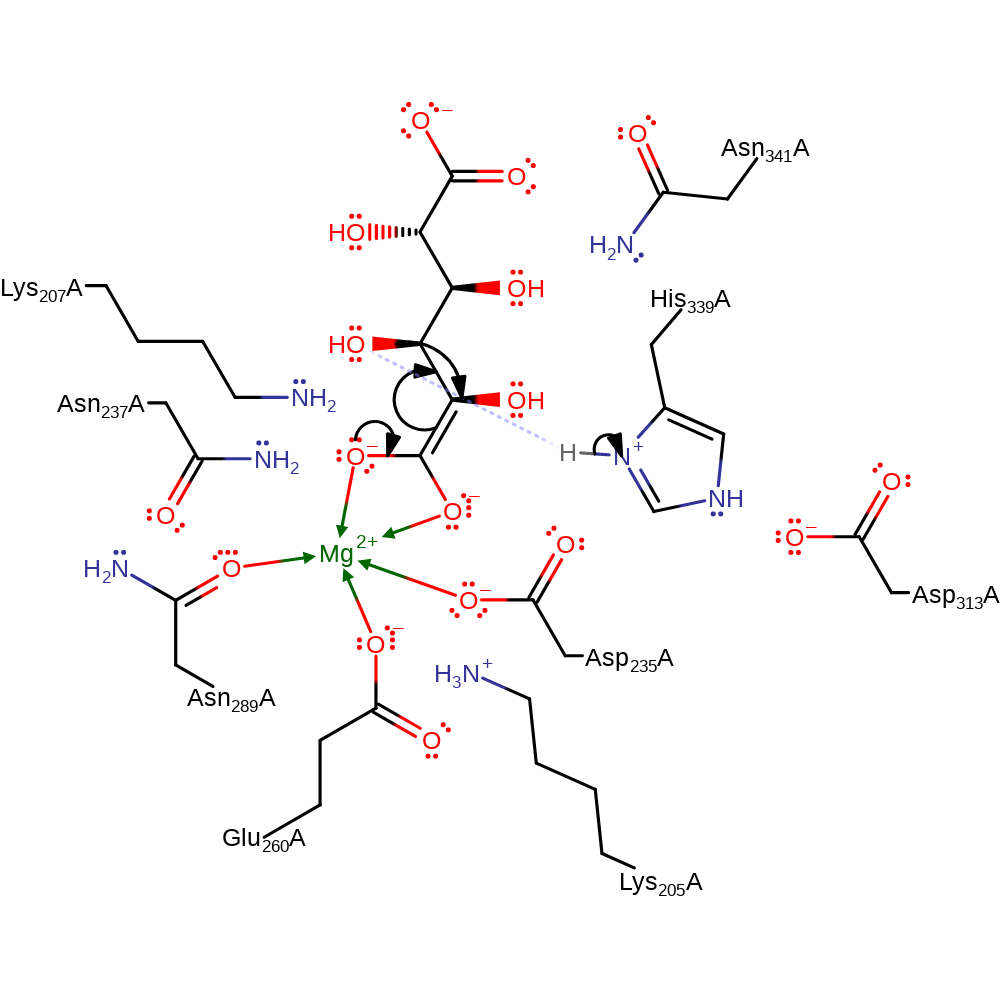
Step 2. The negative intermediate collapses to eliminate water, which abstracts a proton from His339.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu260A | metal ligand |
| Asp235A | metal ligand |
| Asn289A | metal ligand |
| Lys205A | electrostatic stabiliser |
| Asn341A | activator |
| Asp313A | electrostatic stabiliser |
| Asn237A | activator |
| Lys207A | electrostatic stabiliser |
| His339A | proton donor |



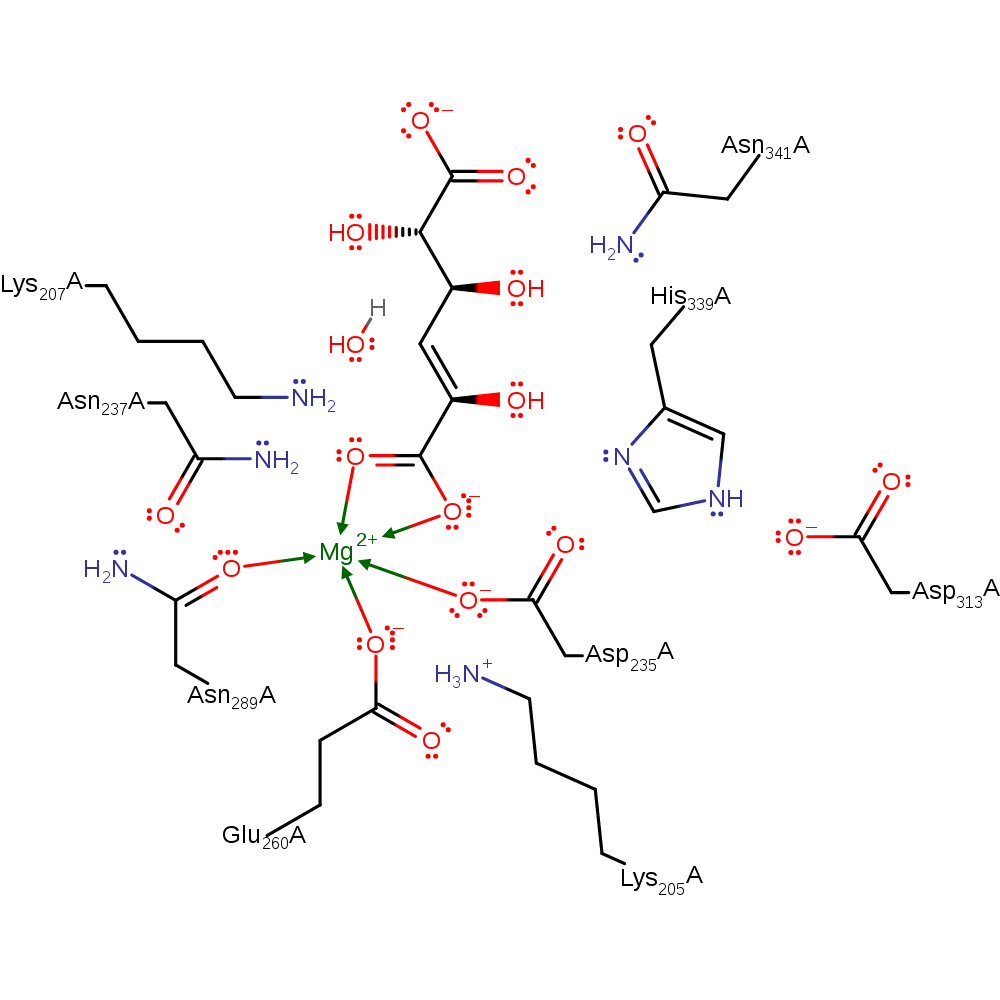 Download:
Download: