N-succinylamino acid racemase
N-succinylamino acid racemase (NAAAR) catalyses the racemisation of N-succinyl-D/L-amino acids, as part of a pathway for the conversion of D- to L-amino acids. NAAARs act on a broad range of N-acylamino acids rather than amino acids. This function is of significant interest in industry where enantiopure amino acids are inportant as chiral building blocks for antibiotics, herbicides, and drugs. The experimentally characterised enzyme from Geobacillus kaustophilus efficiently catalyses the racemisation of hydrophobic, polar, and some basic N-succinyl-D/L-amino acids.
These enzymes are members of the enolase superfamily and highly similar to enzymes in the o-succinylbenzoate synthase family, and some may also function biologically as OSBSs. Although the reaction catalyzed by this family is similar to that catalyzed by the NSAR2 family (differing only in the preference of N-succinyl arginine/lysine for NSAR2 versus N-succinyl hydrophobic amino acids for NSAR), phylogenetic analysis suggests that these two families have independent evolutionary origins within the enolase superfamily. Not surprisingly, the amino acids responsible for substrate recognition appear to differ between the two families.
Reference Protein and Structure
- Sequence
-
Q9RYA6
 (4.2.1.113)
(4.2.1.113)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Deinococcus radiodurans R1 (Bacteria)

- PDB
-
1r0m
- Structure of Deinococcus radiodurans N-acylamino acid racemase at 1.3 : insights into a flexible binding pocket and evolution of enzymatic activity
(1.3 Å)



- Catalytic CATH Domains
-
3.20.20.120
 (see all for 1r0m)
(see all for 1r0m)
- Cofactors
- Magnesium(2+) (1)
Enzyme Mechanism
Introduction
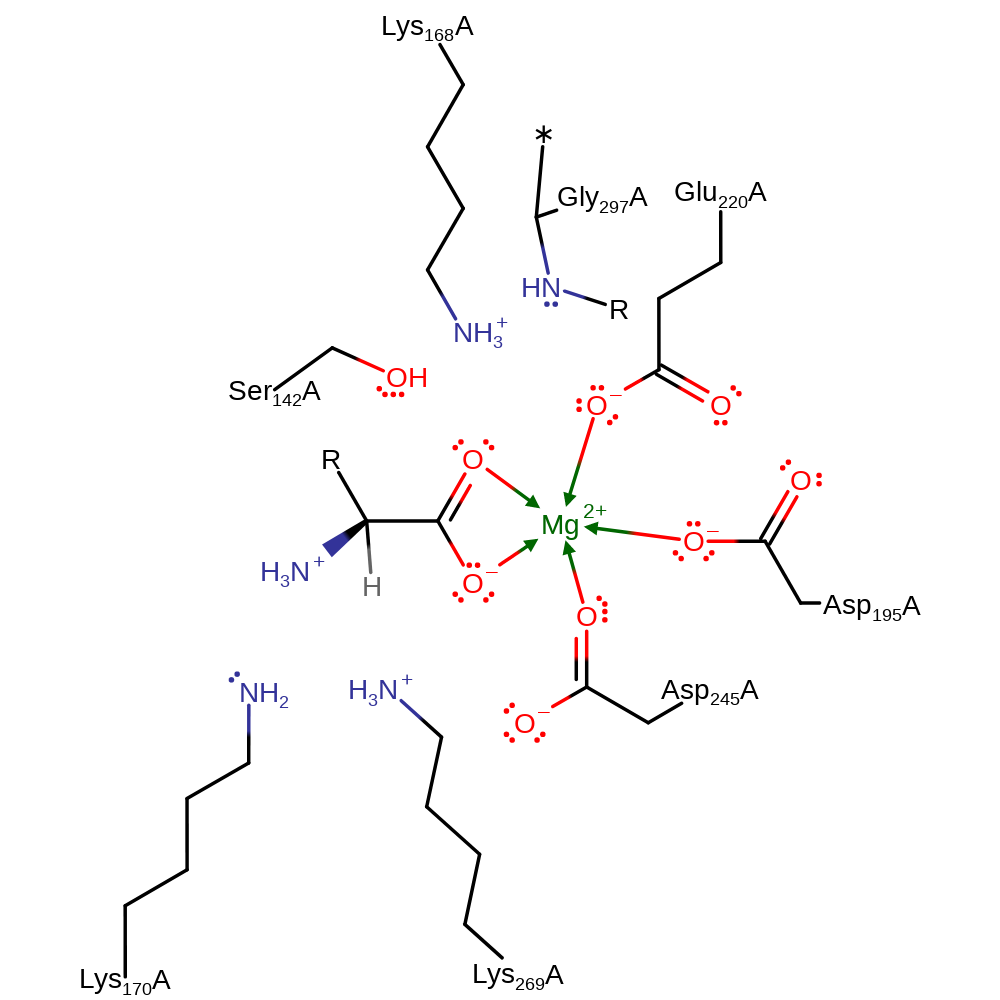
The mechanism is proposed to proceed via a two-base proton abstraction. The alpha-proton of an N-acylamino acid is abstracted by a neutral Lys to produce the enolic intermediate stabilized by the divalent metal ion. A proton is donated by the other (positive) Lys to the intermediate to yield the racemised product.This second Lys can then can then reversibly act as the principle base as shown in the scheme.
Catalytic Residues Roles
| UniProt | PDB* (1r0m) | ||
| Lys170 | Lys170A | General acid/base. In the direction shown here, donates its proton to the substrate to give the product. | proton donor |
| Asp195, Glu220, Asp245 | Asp195A, Glu220A, Asp245A | Bind the Mg(II) ion. | metal ligand |
| Lys269 | Lys269A | Acts as a general acid/base. | proton acceptor |
| Gly297 (main-N), Ser142, Lys168 | Gly297A (main-N), Ser142A, Lys168A | Stabilise the oxyanion intermediate through hydrogen bonding. | transition state stabiliser |
Chemical Components
assisted keto-enol tautomerisation, proton transferReferences
- Chiu WC et al. (2006), J Mol Biol, 359, 741-753. Structure–Stability–Activity Relationship in Covalently Cross-linked N-Carbamoyl d-Amino acid Amidohydrolase and N-Acylamino acid Racemase. DOI:10.1016/j.jmb.2006.03.063. PMID:16650857.
- Soriano-Maldonado P et al. (2015), Mol Biotechnol, 57, 454-465. Biochemical and Mutational Characterization of N-Succinyl-Amino Acid Racemase from Geobacillus stearothermophilus CECT49. DOI:10.1007/s12033-015-9839-4. PMID:25875730.
- Sakai A et al. (2006), Biochemistry, 45, 4455-4462. Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway for the irreversible conversion of D- to L-amino acids. DOI:10.1021/bi060230b. PMID:16584181.
- Wang WC et al. (2004), J Mol Biol, 342, 155-169. Structural Basis for Catalytic Racemization and Substrate Specificity of an N-Acylamino Acid Racemase Homologue from Deinococcus radiodurans. DOI:10.1016/j.jmb.2004.07.023. PMID:15313614.
- Gulick AM et al. (2001), Biochemistry, 40, 15716-15724. Evolution of Enzymatic Activities in the Enolase Superfamily: Crystal Structures of thel-Ala-d/l-Glu Epimerases fromEscherichia coliandBacillus subtilis†,‡. DOI:10.1021/bi011641p.
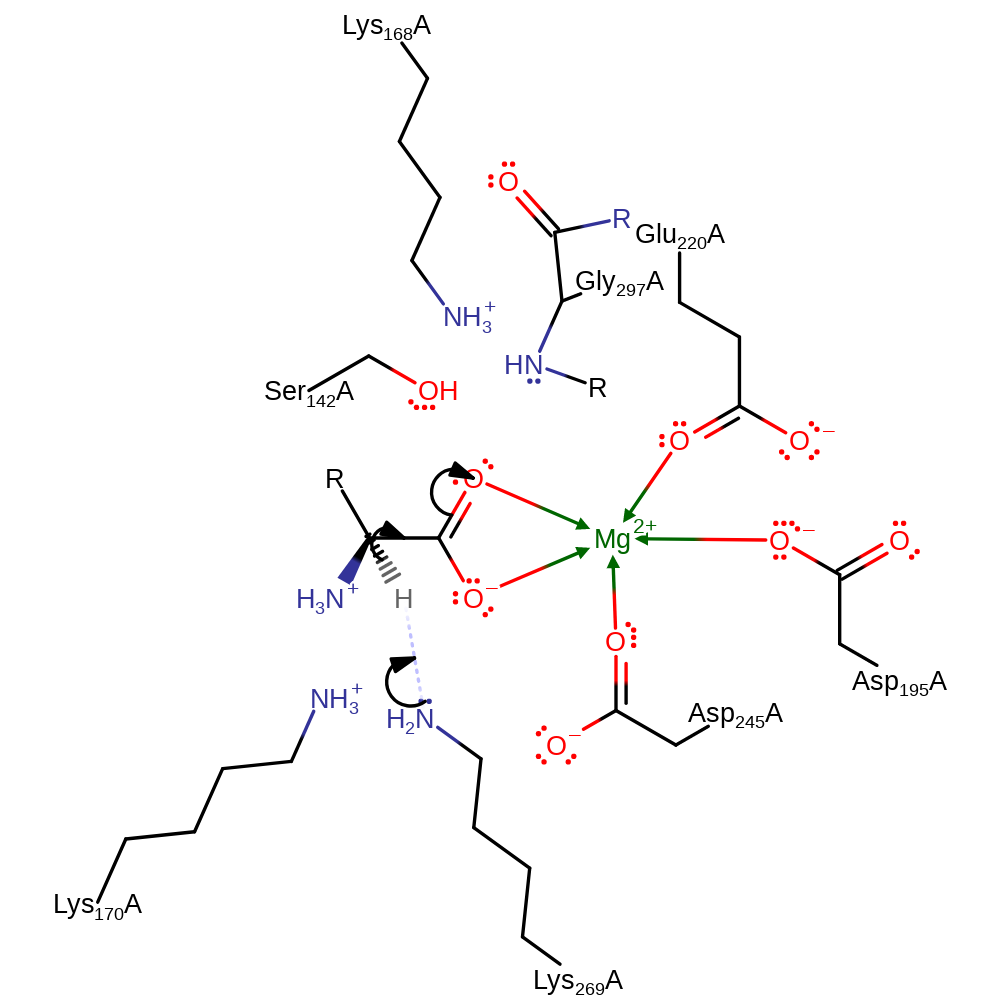
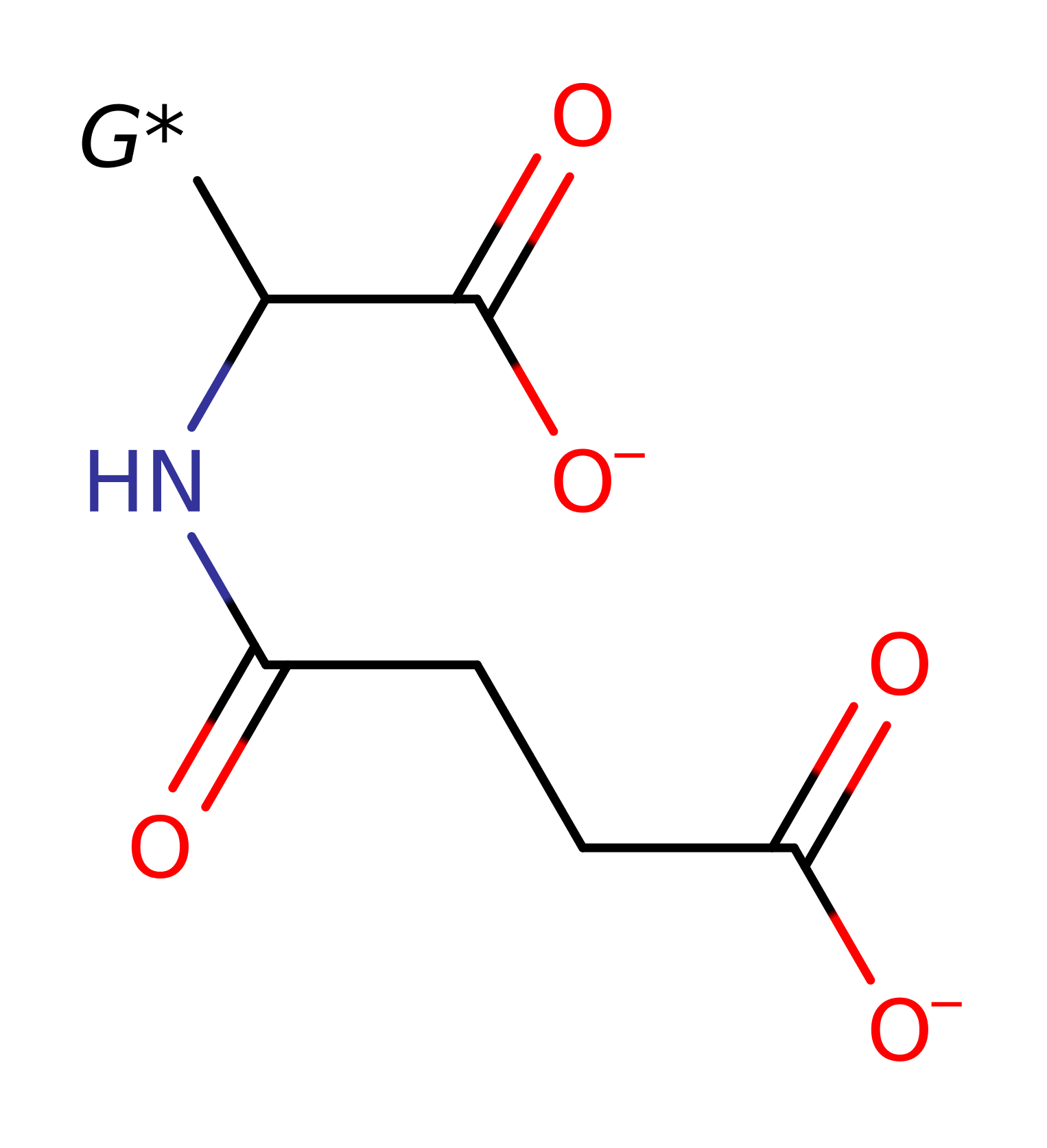
Step 1. Lys269 abstracts a proton from the substrate with concomitant double bond rearrangement.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser142A | transition state stabiliser |
| Lys168A | transition state stabiliser |
| Gly297A (main-N) | transition state stabiliser |
| Glu220A | metal ligand |
| Asp245A | metal ligand |
| Asp195A | metal ligand |
| Lys269A | proton acceptor |
Chemical Components
assisted keto-enol tautomerisation, proton transfer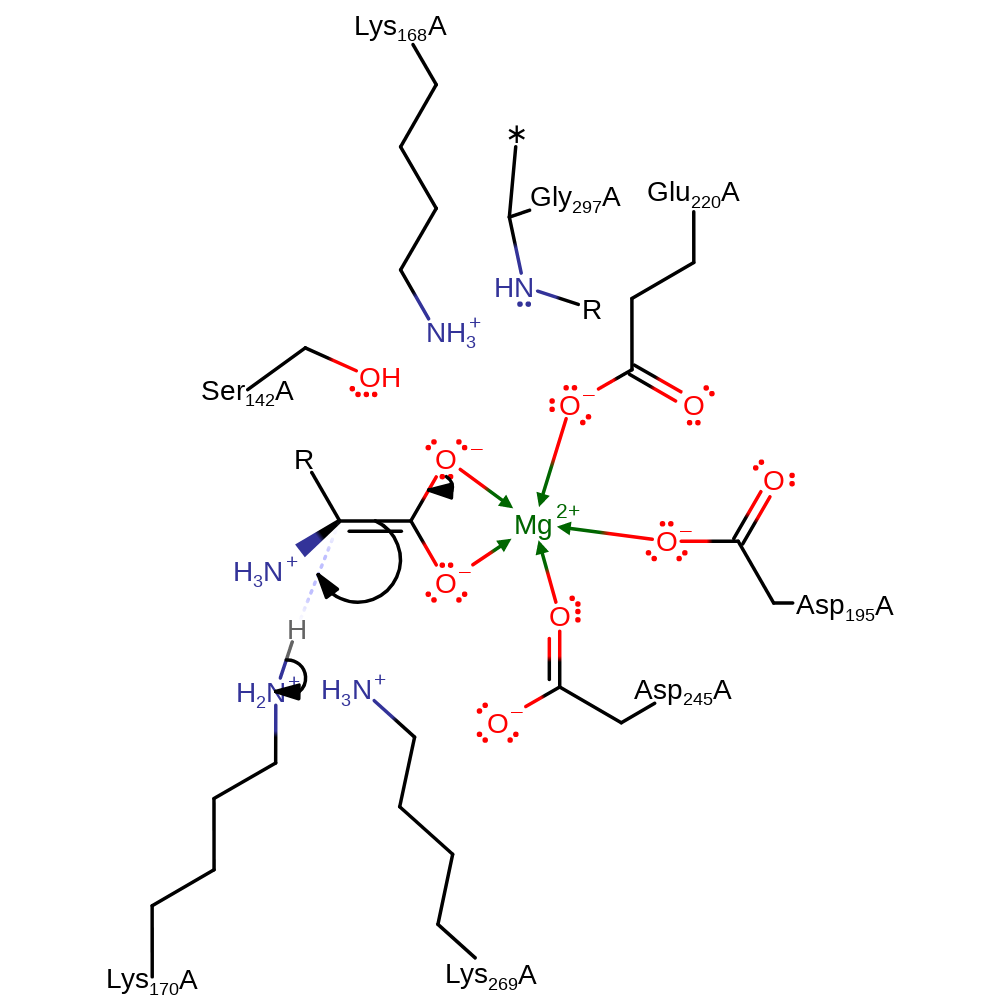
Step 2. The oxyanion intermediate collapses, with concomitant double bond rearrangement and deprotonation of Lys170.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp245A | metal ligand |
| Asp195A | metal ligand |
| Gly297A (main-N) | transition state stabiliser |
| Lys168A | transition state stabiliser |
| Ser142A | transition state stabiliser |
| Lys170A | proton donor |
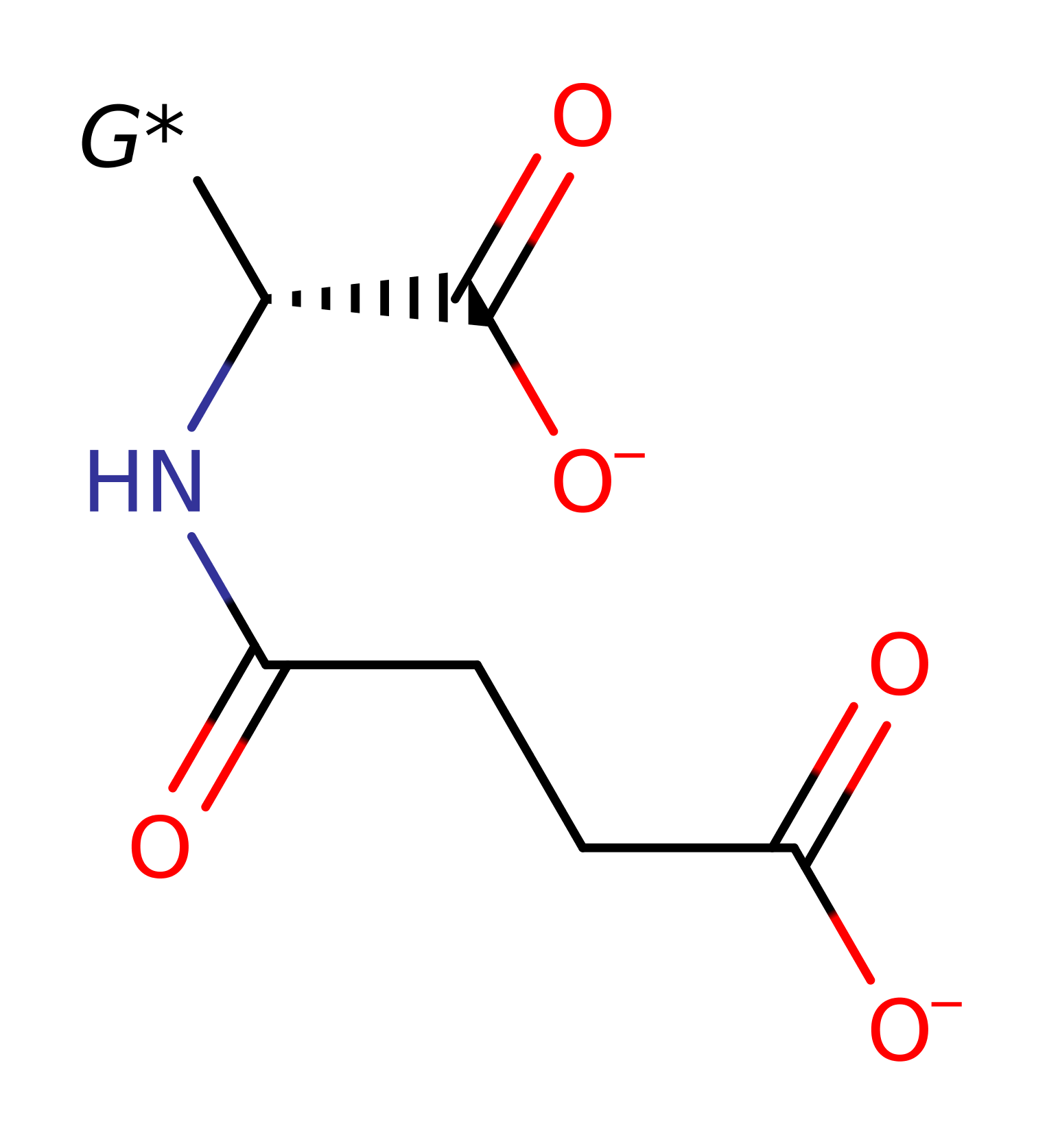
 Download:
Download: