UDP-N-acetylenolpyruvylglucosamine reductase (MurB)
UDP-N-acetylenolpyruvylglucosamine reductase (MurB) reduces both E and Z isomers of enolbutyryl-UDP-GlcBAc analogs of the C3 enolpyruvate substate to UDP-methyl-N-acetylmuramic acid in the presence of NADPH. The overall product of this metabolic pathway, petidoglycan, is a biopolymer unique to Gram-positive and Gram-negative bacteria for which is essential for maintaining osmotic cell wall integrity. The absence of a homologue in eukaryotic cells makes MurB an attractive target for small molecule inhibitors with the potential to have broad antibacterial activity.
The ability of MurB to catalyse the stereo-selective reduction of both E and Z isomers of the substrate is thought to result from the active site architecture restricting free rotation around the C2-C3 bond, and slowing the rate relative to reprotonation. Structural data show the functional groups thought to be involved in the hydride transfer to C3 and protonation at C2 of the enol-ether substrate are arranged anti relative to the enol-double bond. From this information, the stereochemical outcome was predicted to yield a 2R,3R-dideuterio product. This product was later identified using chemical synthetic analysis and comparative NMR.
Reference Protein and Structure
- Sequence
-
P08373
 (1.3.1.98)
(1.3.1.98)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1mbb
- OXIDOREDUCTASE
(2.3 Å)



- Catalytic CATH Domains
-
3.30.465.10
 3.90.78.10
3.90.78.10  (see all for 1mbb)
(see all for 1mbb)
- Cofactors
- Fadh2(2-) (1), Potassium(1+) (1)
Enzyme Reaction (EC:1.3.1.98)
Enzyme Mechanism
Introduction
The mechanism proceeds as follows: NADPH binds to the enzyme and hydride transfer of the 4-pro -S hydrogen of NADPH to N5 of an enzyme-bound flavin (FAD cofactor). Release of NADP+ is followed by the binding of UDP-GlcNAc to the enzyme active site. Hydride transfer from the reduced flavin to C3 of the enolpyruvyl moiety of the UDP-sugar substrate generates a carbanion equivalent at C2, which can be stabilised by the R -carboxylate at C1 as an enol intermediate. A proton from Ser229 is then transferred to C2, giving the UDPMurNAc product.
Catalytic Residues Roles
| UniProt | PDB* (1mbb) | ||
| Ser229 | Ser229A | Acts as a general acid to quench the enol intermediate to form the acid product. The acid/base role proposed for Ser229 is unusual. The pKa of a serine residue hydroxyl group is typically in the range of 13-15, making proton donation at neutral pH energetically unfavourable. However, in this reaction where Ser229 acts as an electrophile, it is a comparatively favourable proton donor, it is located and orientated to perform this function and loss of function mutagenesis studies indicate Ser299 to be a key catalytic residue. | activator, proton acceptor, proton donor |
| Arg159, Glu325 | Arg159A, Glu325A | Acts to stabilise carbanion intermediate as an enol by hydrogen-bonding. | attractive charge-charge interaction, electrostatic stabiliser, increase electrophilicity |
Chemical Components
hydride transfer, cofactor used, native state of cofactor is not regenerated, proton transfer, native state of cofactor regenerated, intermediate formation, overall reactant used, intermediate collapse, overall product formed, native state of enzyme regenerated, inferred reaction stepReferences
- Benson TE et al. (1997), Biochemistry, 36, 796-805. Kinetic Characterization of Wild-Type and S229A Mutant MurB: Evidence for the Role of Ser 229 as a General Acid†. DOI:10.1021/bi962220o. PMID:9020777.
- Nishida S et al. (2006), J Biol Chem, 281, 1714-1724. Identification and characterization of amino acid residues essential for the active site of UDP-N-acetylenolpyruvylglucosamine reductase (MurB) from Staphylococcus aureus. DOI:10.1074/jbc.M509277200. PMID:16236703.
- Constantine KL et al. (1997), J Mol Biol, 267, 1223-1246. Characterization of NADP+ binding to perdeuterated MurB: backbone atom NMR assignments and chemical-shift changes. DOI:10.1006/jmbi.1997.0915. PMID:9150408.
- Lees WJ et al. (1996), Biochemistry, 35, 1342-1351. (E)-Enolbutyryl-UDP-N-acetylglucosamine as a Mechanistic Probe of UDP-N-acetylenolpyruvylglucosamine Reductase (MurB)†,‡. DOI:10.1021/bi952287w. PMID:8634262.

Step 1. NADPH binds in close proximity to Ser229 and transfers a hydrogen to the N5 of the enzyme bound flavin cofactor. It is uncertain which residues are specifically involved in promoting the hydride transfer from the NADPH cofactor to the enzyme bound flavin. NMR analysis indicates NADPH to bind in the vicinity of Ser229, as well as several aromatic residues [PMID:9020777]. A monovalent cation is required for catalysis, although no metal is present in the crystal structure.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
hydride transfer, cofactor used, native state of cofactor is not regenerated
Step 2. The flavin cofactor transfers a hydride atom from N5 to the re-face of the C2-C3 enolpyruvyl group. Either Glu325 or Arg159 could act as the proton donor: both are correctly positioned and orientated within the active site, and both are required for catalysis [PMID:8634262].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu325A | attractive charge-charge interaction, increase electrophilicity, electrostatic stabiliser |
| Arg159A | attractive charge-charge interaction, electrostatic stabiliser, increase electrophilicity |
| Glu325A | proton donor |
Chemical Components
hydride transfer, proton transfer, native state of cofactor regenerated, intermediate formation, overall reactant used
Step 3. Glu325 acts as a general base, taking back the enol proton. This initiates the conjugate, stereo-selective proton abstraction by the enolate from Ser229. For MerB to catalyse the stereoselective reduction of both E and Z substrate isomers, the rate of rotation of the C2-C3 bond must be slow relative to reprotonation. The active site architecture ensures stereoselective discrimination in the formation of the enantio-selective product [PMID:8634262].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu325A | activator, attractive charge-charge interaction, electrostatic stabiliser, increase basicity |
| Arg159A | attractive charge-charge interaction, electrostatic stabiliser |
| Ser229A | activator |
| Ser229A | proton donor |
| Glu325A | proton acceptor |
Chemical Components
proton transfer, intermediate collapse, overall product formed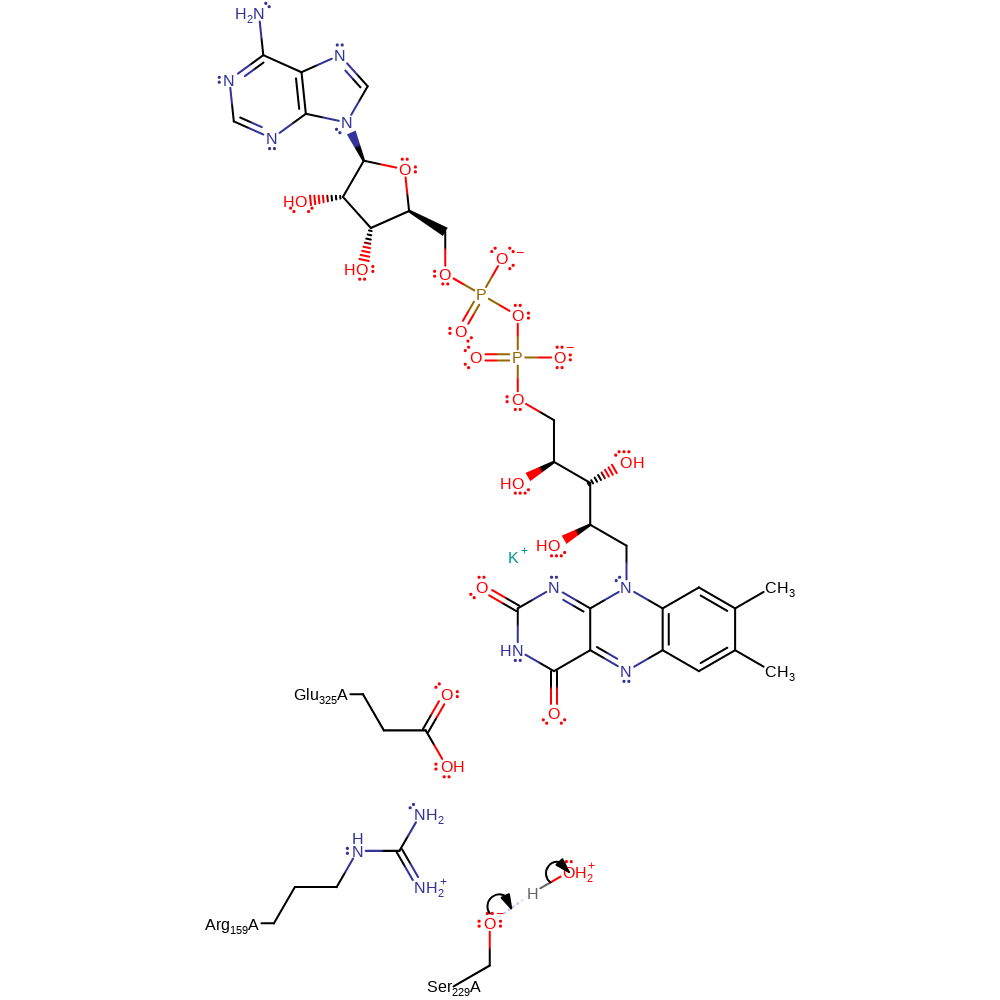
Step 4. Reprotonation of Ser229 regenerates the active site. Proton labelling studies indicate that Ser229 is exposed to solvent, and so it is inferred that the residue is regenerated by reprotonation from a solvent molecule [PMID:8634262].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser229A | proton acceptor |





 Download:
Download: