Galactose oxidiase
Galactose oxidase is a copper containing enzyme which oxidises primary alcohols to aldehydes by the reduction of molecular oxygen to hydrogen peroxide. The enzyme catalyses the oxidation of many substrates, including dihydroxyacetone, small molecules and olysaccharides by use of a protein radical cofactor, a novel metalloradical complex, formed by a unique structural feature at the copper site with a novel thioether bond linking Cys 228 and Tyr 272 formed from a post-translational modification, which is in a stacking interaction with Trp 290. Galactose oxidase is remarkable in the extent to which free radicals are involved in all aspects of the enzyme function: serving as a key feature of the active site structure, defining the characteristic reactivity of the complex, and directing the biogenesis of the Tyr-Cys cofactor during protein maturation.
Reference Protein and Structure
- Sequence
-
P0CS93
 (1.1.3.9)
(1.1.3.9)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Fusarium graminearum (Fungus)

- PDB
-
1gog
- NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE
(1.9 Å)



- Catalytic CATH Domains
-
2.130.10.80
 2.60.40.10
2.60.40.10  (see all for 1gog)
(see all for 1gog)
- Cofactors
- Copper(2+) (1) Metal MACiE
Enzyme Reaction (EC:1.1.3.9)
Enzyme Mechanism
Introduction
In the first (reductive) half-reaction, the oxidized free radicals Cu2+ complex reacts with the primary alcohol. The initial proton abstraction from a coordinated substrate activates the alcohol toward inner sphere electron transfer to the Cu(II) metal centre in an unfavourable redox equilibrium, forming an alkoxy radical which undergoes hydrogen atom abstraction by the tyrosine-cysteine phenoxyl free radical ligand to form the product aldehyde. In the second (oxidative) half-reaction, the reduced enzyme reacts with dioxygen , converting the active site to the metalloradical complex and forming hydrogen peroxide. A two-electron oxidation performed by a single copper ion- is explained by the involvement of another redox center, namely a tyrosyl radical from the protein, during the catalytic cycle. Extensive characterizations have shown, that it is located at Tyr272.
Catalytic Residues Roles
| UniProt | PDB* (1gog) | ||
| Cys269 | Cys228A | Forms a thioether cross link with Tyr272- see the functional notes for more details on its action. | covalently attached, activator, metal ligand |
| Tyr313 | Tyr272A | The Try272 forms a cross link to Cys228, which coordinates to copper to form a metalloradical complex in the oxidised, active form of the enzyme This free radical-coupled copper complex is extremely stable. It reacts readily with a variety of electron donors, undergoing single-electron reduction to form a catalytically inactive non-radical Cu(II) complex. Further reduction converts the Cu(II) centre to Cu(I), forming a fully reduced complex that is competent for reaction with O2 and represents a catalytic intermediate in the reaction. The reactivity of the Cys-Tyr cofactor may be reflected in its lower O- H bond dissociation energy in its reduced form, facilitating hydrogen atom donation to O2 during catalysis The orientation of Tyr272 also maximises overlap between the redox orbital on the phenoxyl oxygen and the half-occupied d-orbital on Cu, reflected in a strong antiferromagnetic exchange splitting in the electronic ground state of the complex. | hydrogen radical acceptor, hydrogen radical donor, activator, metal ligand |
| Trp331 | Trp290A | The active site tryptophan (Trp290), which lies over the Tyr-Cys cofactor with the indole nitrogen adjacent to the coordinated oxygen of the Tyr272 side chain may play a role in the stabilisation of the substrate-derived proton in this complex. It is pi-stacked to Try-Cys moiety and also controls entry to the active site. | activator, radical stabiliser |
| Tyr536 | Tyr495A | Tyr495 serves as a general base that activates the reducing substrate by proton abstraction during the reductive half-reaction, and facilitates displacement of hydrogen peroxide product by protonation in the oxidative half-reaction. | activator, metal ligand, proton acceptor, proton donor |
| His537, His622 | His496A, His581A | Form part of the copper binding site. | metal ligand |
Chemical Components
proton transfer, hydrogen transfer, electron transfer, overall product formed, cofactor used, coordination, coordination to a metal ionReferences
- Humphreys KJ et al. (2009), J Am Chem Soc, 131, 4657-4663. Galactose Oxidase as a Model for Reactivity at a Copper Superoxide Center. DOI:10.1021/ja807963e. PMID:19290629.
- Cowley RE et al. (2016), J Am Chem Soc, 138, 13219-13229. Structure of the Reduced Copper Active Site in Preprocessed Galactose Oxidase: Ligand Tuning for One-Electron O2Activation in Cofactor Biogenesis. DOI:10.1021/jacs.6b05792. PMID:27626829.
- Rokhsana D et al. (2012), Inorg Chem, 51, 3513-3524. Role of the Tyr-Cys Cross-link to the Active Site Properties of Galactose Oxidase. DOI:10.1021/ic2022769. PMID:22372371.
- Rannes JB et al. (2011), J Am Chem Soc, 133, 8436-8439. Glycoprotein Labeling Using Engineered Variants of Galactose Oxidase Obtained by Directed Evolution. DOI:10.1021/ja2018477. PMID:21526835.
- Lee YK et al. (2008), Biochemistry, 47, 6637-6649. The Electronic Structure of the Cys-Tyr•Free Radical in Galactose Oxidase Determined by EPR Spectroscopy†. DOI:10.1021/bi800305d. PMID:18512952.
- Rogers MS et al. (2008), Biochemistry, 47, 10428-10439. Cross-Link Formation of the Cysteine 228−Tyrosine 272 Catalytic Cofactor of Galactose Oxidase Does Not Require Dioxygen†‡. DOI:10.1021/bi8010835. PMID:18771294.
- Rogers MS et al. (2007), Biochemistry, 46, 4606-4618. The Stacking Tryptophan of Galactose Oxidase: A Second-Coordination Sphere Residue that Has Profound Effects on Tyrosyl Radical Behavior and Enzyme Catalysis†,‡. DOI:10.1021/bi062139d. PMID:17385891.
- Wilkinson D et al. (2004), Protein Eng Des Sel, 17, 141-148. Structural and kinetic studies of a series of mutants of galactose oxidase identified by directed evolution. DOI:10.1093/protein/gzh018. PMID:15047910.
- Deacon SE et al. (2004), Chembiochem, 5, 972-979. Enhanced Fructose Oxidase Activity in a Galactose Oxidase Variant. DOI:10.1002/cbic.200300810. PMID:15239055.
- Whittaker JW (2003), Chem Rev, 103, 2347-2364. Free Radical Catalysis by Galactose Oxidase. DOI:10.1021/cr020425z. PMID:12797833.
- Whittaker JW (2002), Adv Protein Chem, 60, 1-49. Galactose oxidase. PMID:12418174.
- Thomas F et al. (2002), Angew Chem Int Ed Engl, 41, 3047-3050. A Structural and Functional Model of Galactose Oxidase: Control of the One-Electron Oxidized Active Form through Two Differentiated Phenolic Arms in a Tripodal Ligand. DOI:10.1002/1521-3773(20020816)41:16<3047::aid-anie3047>3.0.co;2-w. PMID:12203454.
- Firbank SJ et al. (2001), Proc Natl Acad Sci U S A, 98, 12932-12937. Crystal structure of the precursor of galactose oxidase: An unusual self-processing enzyme. DOI:10.1073/pnas.231463798. PMID:11698678.
- Himo F et al. (2000), J Am Chem Soc, 122, 8031-8036. Catalytic Mechanism of Galactose Oxidase: A Theoretical Study. DOI:10.1021/ja994527r.
- Whittaker MM et al. (2000), J Mol Catal B Enzym, 8, 3-15. Spectroscopic and magnetochemical studies on the active site copper complex in galactose oxidase. DOI:10.1016/s1381-1177(99)00072-7.
- Rothlisberger U et al. (2000), J Biol Inorg Chem, 5, 236-250. A comparative study of galactose oxidase and active site analogs based on QM/MM Car-Parrinello simulations. PMID:10819469.
- Whittaker MM et al. (1998), Biochemistry, 37, 8426-8436. Kinetic Isotope Effects as Probes of the Mechanism of Galactose Oxidase†. DOI:10.1021/bi980328t. PMID:9622494.
- Itoh S et al. (1997), Inorg Chem, 36, 1407-1416. Active Site Models for Galactose Oxidase. Electronic Effect of the Thioether Group in the Novel Organic Cofactor. DOI:10.1021/ic961144a.
- Reynolds MP et al. (1997), J Biol Inorg Chem, 2, 327-335. Structure and mechanism of galactose oxidase: catalytic role of tyrosine 495. DOI:10.1007/s007750050139.
- Wachter RM et al. (1996), Biochemistry, 35, 14425-14435. Thiols as Mechanistic Probes for Catalysis by the Free Radical Enzyme Galactose Oxidase†. DOI:10.1021/bi961369x. PMID:8916929.
- Reynolds MP et al. (1995), Biochem Soc Trans, 23, 510S-. Tyrosine 495 is a key residue in the active site of galactose oxidase. PMID:8654695.
- Ito N et al. (1994), J Mol Biol, 238, 704-814. Crystal Structure of a Free Radical Enzyme, Galactose Oxidase. DOI:10.1006/jmbi.1994.1335. PMID:8182749.
- Baron AJ et al. (1994), J Biol Chem, 269, 25095-25105. Structure and mechanism of galactose oxidase. The free radical site. PMID:7929198.
- Whittaker MM et al. (1993), Biophys J, 64, 762-772. Ligand interactions with galactose oxidase: mechanistic insights. DOI:10.1016/s0006-3495(93)81437-1. PMID:8386015.
- Ito N et al. (1991), Nature, 350, 87-90. Novel thioether bond revealed by a 1.7 Å crystal structure of galactose oxidase. DOI:10.1038/350087a0. PMID:2002850.
- Whittaker MM et al. (1988), J Biol Chem, 263, 6074-6080. The active site of galactose oxidase. PMID:2834363.

Step 1. The coordinated, anionic Tyr residue abstracts a proton from the coordinated primary alcohol substrate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr495A | activator |
| Tyr272A | metal ligand |
| Cys228A | covalently attached |
| Trp290A | radical stabiliser, activator |
| Tyr495A | proton acceptor |
Chemical Components
proton transferCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr272A | activator |
| Cys228A | covalently attached |
| Trp290A | radical stabiliser |
| His581A | metal ligand |
| His496A | metal ligand |
| Tyr495A | metal ligand |
| Tyr272A | metal ligand, hydrogen radical acceptor |
Chemical Components
hydrogen transfer
Step 3. One electron is transferred from the radical-anion alcohol to the Cu centre, reducing the metal from +2 to +1
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr272A | metal ligand |
| Cys228A | covalently attached |
| Trp290A | radical stabiliser |
| His581A | metal ligand |
| His496A | metal ligand |
| Tyr495A | metal ligand |
Chemical Components
electron transfer, overall product formed, cofactor used
Step 4. Dioxygen forms a superoxide-radial anion on withdrawing an electron from the Cu centre, oxidising the metal to the +2 state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr272A | metal ligand |
| His581A | metal ligand |
| His496A | metal ligand |
| Tyr495A | metal ligand |
| Cys228A | covalently attached |
Chemical Components
coordination, electron transfer, coordination to a metal ion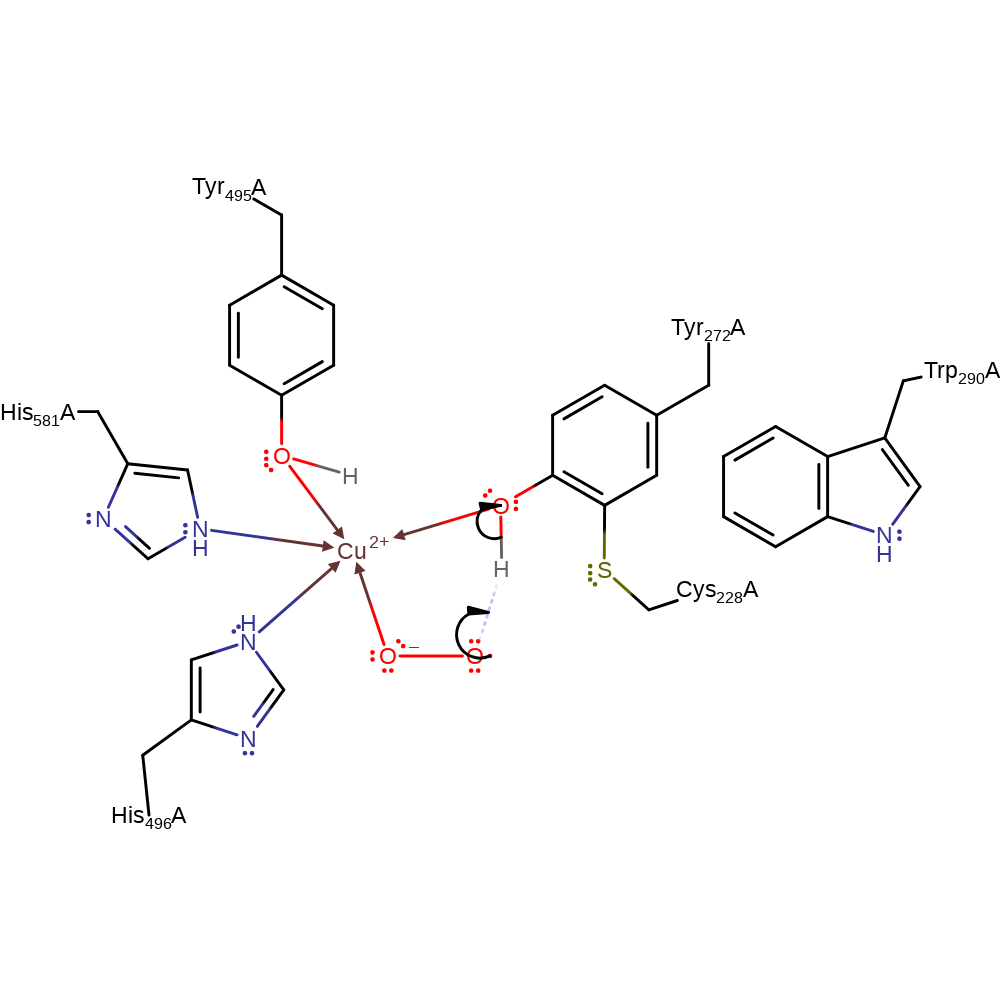
Step 5. The oxide radical initiates homolytic hydrogen atom abstraction at Tyr272, regenerating the radical tyrosine ligand.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His581A | metal ligand |
| Tyr272A | activator |
| Cys228A | covalently attached |
| His496A | metal ligand |
| Tyr495A | metal ligand |
| Cys228A | activator |
| Trp290A | radical stabiliser |
| Tyr272A | hydrogen radical donor |
Chemical Components
hydrogen transfer
Step 6. The anionic peroxide abstracts a proton from Tyr495. After this step has occurred, hydrogen peroxide is substituted from the Cu(II) coordination sphere by water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr272A | metal ligand |
| Trp290A | radical stabiliser |
| Cys228A | covalently attached |
| His581A | metal ligand |
| His496A | metal ligand |
| Tyr495A | metal ligand |
| Cys228A | metal ligand |
| Tyr495A | proton donor |





 Download:
Download: