Argininosuccinate synthetase
Argininosuccinate synthase (EC:6.3.4.5) (AS) is a urea cycle enzyme that catalyzes the penultimate (seventh) step in arginine biosynthesis: the condensation of glutamate and citrulline to form agininosuccinate, AMP and diphosphate.
In humans, a defect in the AS gene causes citrullinemia, a genetic disease characterised by severe vomiting spells and mental retardation [PMID:19006241].
AS is a homotetrameric enzyme of chains of about 400 amino-acid residues. An arginine seems to be important for the enzyme's catalytic mechanism. The sequences of AS from various prokaryotes, archaebacteria and eukaryotes show significant similarity.
Although there are two sub-types of argininosuccinate synthase, they appear to have the same mechanism and so are both represented by this entry.
Reference Protein and Structure
- Sequence
-
P59846
 (6.3.4.5)
(6.3.4.5)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Thermus thermophilus HB8 (Bacteria)

- PDB
-
1j21
- Crystal Structure of Thermus thermophilus HB8 Argininosuccinate Synthetase in complex with ATP and citrulline
(2.2 Å)



- Catalytic CATH Domains
-
3.40.50.620
 3.90.1260.10
3.90.1260.10  (see all for 1j21)
(see all for 1j21)
- Cofactors
- Magnesium(2+) (1) Metal MACiE
Enzyme Reaction (EC:6.3.4.5)
Enzyme Mechanism
Introduction
The catalytic reaction has been proposed to be initiated by the nucleophilic attack of the citrulline ureido oxygen on the alpha-phopshate of ATP, followed by the formation of a citrullyl-AMP intermediate, if aspartate is bound at this stage, the kinetics of the step are stimulated by a factor of 600. This activated citrulline AMP intermediate is formed via a pentavalent phosphate intermediate stabilised by Arg 106. The alpha-amino group of L-aspartate then attacks the imino carbon atom of the citrullyl-AMP intermediate, producing AMP and argininosuccinate.
Catalytic Residues Roles
| UniProt | PDB* (1j21) | ||
| Arg92 | Arg92A | Acts to stabilise the negatively charged pentavalent transition states formed during the course of the reaction. | hydrogen bond donor, electrostatic stabiliser |
| Asp121 | Asp121A | Deprotonates the aspartate substrate amino group to allow the nitrogen atom to act as a nucleophile. | proton acceptor, proton donor |
| Ser173, Asp12 | Ser173A, Asp12A | Act to hold the ATP in the correct 'S' conformation so that the reaction can occur. | hydrogen bond donor, steric role |
Chemical Components
bimolecular nucleophilic substitution, proton transfer, bimolecular nucleophilic addition, unimolecular elimination by the conjugate base, native state of enzyme regeneratedReferences
- Goto M et al. (2003), J Biol Chem, 278, 22964-22971. Structures of Argininosuccinate Synthetase in Enzyme-ATP Substrates and Enzyme-AMP Product Forms: STEREOCHEMISTRY OF THE CATALYTIC REACTION. DOI:10.1074/jbc.m213198200. PMID:12684518.
- Engel K et al. (2009), Hum Mutat, 30, 300-307. Mutations and polymorphisms in the human argininosuccinate synthetase (ASS1) gene. DOI:10.1002/humu.20847. PMID:19006241.
- Karlberg T et al. (2008), Acta Crystallogr D Biol Crystallogr, 64, 279-286. Structure of human argininosuccinate synthetase. DOI:10.1107/s0907444907067455. PMID:18323623.
- Curis E et al. (2005), Amino Acids, 29, 177-205. Almost all about citrulline in mammals. DOI:10.1007/s00726-005-0235-4. PMID:16082501.
- Lemke CT et al. (2002), J Biol Chem, 277, 13074-13081. Substrate Induced Conformational Changes in Argininosuccinate Synthetase. DOI:10.1074/jbc.m112436200. PMID:11809762.
- Goto M et al. (2002), J Biol Chem, 277, 15890-15896. Crystal Structure of Argininosuccinate Synthetase from Thermus thermophilus HB8. STRUCTURAL BASIS FOR THE CATALYTIC ACTION. DOI:10.1074/jbc.m112430200. PMID:11844799.
- Lemke CT et al. (2001), Structure, 9, 1153-1164. The 1.6 Å Crystal Structure of E. coli Argininosuccinate Synthetase Suggests a Conformational Change during Catalysis. DOI:10.1016/s0969-2126(01)00683-9. PMID:11738042.
- Hilscher LW et al. (1985), Biochemistry, 24, 5888-5893. Measurement of positional isotope exchange rates in enzyme-catalyzed reactions by fast atom bombardment mass spectrometry: application to argininosuccinate synthetase. DOI:10.1021/bi00342a030. PMID:2867775.

Step 1. Citrulline attacks at the alpha phosphate of ATP, which is held in a reactive S conformation by surrounding residues.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg92A | hydrogen bond donor, electrostatic stabiliser |
| Ser173A | hydrogen bond donor, steric role |
| Asp12A | steric role |
Chemical Components
ingold: bimolecular nucleophilic substitution
Step 2. The amine group of aspartate is deprotonated by Asp121, activating the nitrogen towards nucleophilic attack at the AMP-imine adduct. This results in the formation of a tetrahedral intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg92A | hydrogen bond donor |
| Ser173A | hydrogen bond donor |
| Asp121A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition
Step 3. Asp121A is deprotonated by the unprotonated amine group of the tetrahedral intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg92A | hydrogen bond donor |
| Ser173A | hydrogen bond donor |
| Asp121A | proton donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition
Step 4. The tetrahedral intermediate collapses, generating protonated AMP and arginosuccinate - a biosynthetic precursor of arginine.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg92A | hydrogen bond donor |
| Ser173A | hydrogen bond donor |

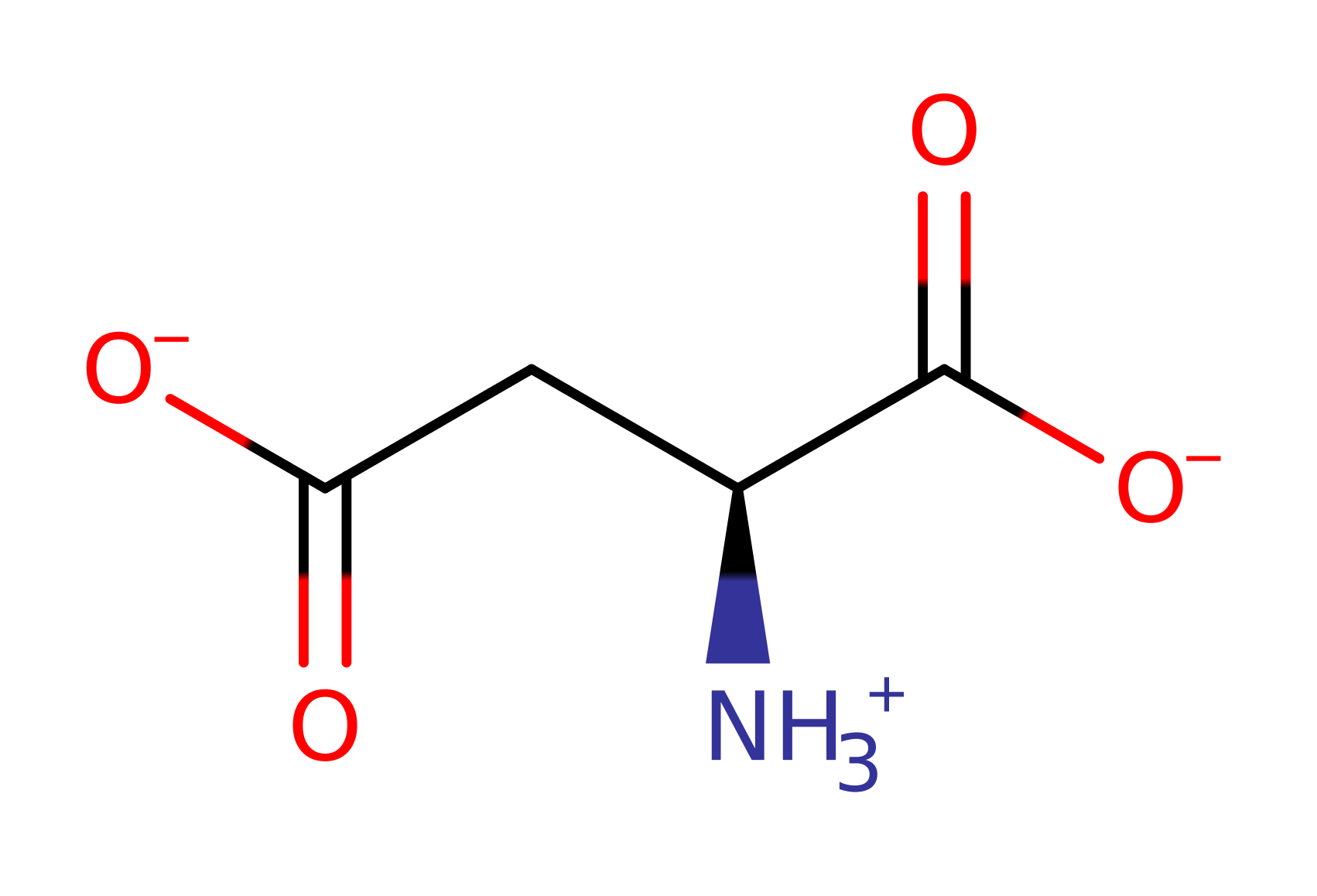





 Download:
Download: