Asparagine synthase (glutamine-hydrolysing)
Asparagine synthetase B catalyses the ATP dependent formation of asparagine from aspartate using either glutamine or ammonia as a nitrogen source. The enzyme is a homodimer with two active sites [PMID:10587437] in each subunit, one responsible for glutamine hydrolysis and one for asparagine synthesis, separated by a hydrophobic intramolecular tunnel around 20A long [PMID:12706338]. Sequencing analysis has shown the synthetase to belong to the larger glutamine transferase family.
Reference Protein and Structure
- Sequence
-
P22106
 (6.3.5.4)
(6.3.5.4)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1ct9
- CRYSTAL STRUCTURE OF ASPARAGINE SYNTHETASE B FROM ESCHERICHIA COLI
(2.0 Å)



- Catalytic CATH Domains
-
3.60.20.10
 3.40.50.620
3.40.50.620  (see all for 1ct9)
(see all for 1ct9)
- Cofactors
- Magnesium(2+) (1) Metal MACiE
Enzyme Reaction (EC:6.3.5.4)
Enzyme Mechanism
Introduction
The enzyme catalyses three distinct chemical reactions: glutamine hydrolysis to yield ammonia (which is then channelled to the second active site) takes place in the N-terminal domain. The C-terminal active site mediates both the synthesis of a beta-aspartyl-AMP intermediate and its subsequent reaction with ammonia. However, the exact order of the partial reactions is still somewhat unclear [PMID:9748330, PMID:12706338], here we show the hydrolysis of the glutamate occurring first.
Glutamine hydrolysis occurs at the N terminal. A nucleophilic Cys residue attacks the glutamine substrate, displacing ammonia, forming a substrate-enzyme intermediate which is then hydrolysed. The ammonia diffuses though the interdomain tunnel from the site of production to the site of utilisation: the synthetase component in the C terminal.
A two site ping pong mechanism for the reaction between the aspartic acid, ATP and ammonia has been implicated with several residues thought to be responsible for the binding the substrates through hydrogen bonding interactions.
Kinetic analysis has shown the rate of catalysis at each site to be independent of one another, and so the stoichiometry between the sites must be maintained by the catalytic efficiency of the two sites.
Catalytic Residues Roles
| UniProt | PDB* (1ct9) | ||
| Cys2 (N-term) | Ala1A (N-term) | Acts as a general acid/base to activate the cysteine nucleophile. | proton acceptor, proton donor |
| Leu51 (main-C) | Leu50A (main-C) | Helps stabilise the reactive intermediates formed. | hydrogen bond acceptor, electrostatic stabiliser |
| Thr322, Arg325 | Thr321A, Arg324A | Bind and stabilise the phosphate groups of the ATP and reactive intermediates formed. | hydrogen bond donor, electrostatic stabiliser |
| Cys2 | Ala1A | Acts as a catalytic nucleophile in the glutaminase domain reaction. | covalently attached, hydrogen bond acceptor, nucleofuge, nucleophile, proton acceptor, proton donor |
| Gly76 (main-N), Asn75 | Gly75A (main-N), Asn74A | Forms the oxyanion hole that stabilises the reactive intermediates and transition states formed. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic addition, proton relay, enzyme-substrate complex formation, overall reactant used, intermediate formation, unimolecular elimination by the conjugate base, enzyme-substrate complex cleavage, deamination, intermediate collapse, overall product formed, native state of enzyme regeneratedReferences
- Tesson AR et al. (2003), Arch Biochem Biophys, 413, 23-31. Revisiting the steady state kinetic mechanism of glutamine-dependent asparagine synthetase from Escherichia coli. DOI:10.1016/s0003-9861(03)00118-8. PMID:12706338.
- Fresquet V et al. (2004), Bioorg Chem, 32, 63-75. Kinetic mechanism of asparagine synthetase from Vibrio cholerae. DOI:10.1016/j.bioorg.2003.10.002. PMID:14990305.
- Larsen TM et al. (2000), Biochemistry, 39, 7330-. Three-dimensional structure of escherichia coli asparagine synthetase B: A short journey from substrate to product. PMID:10852734.
- Larsen TM et al. (1999), Biochemistry, 39, 7330-7330. Three-Dimensional Structure ofEscherichia coliAsparagine Synthetase B: A Short Journey from Substrate to Product. DOI:10.1021/bi005109y. PMID:10587437.
- Boehlein SK et al. (1998), Biochemistry, 37, 13230-13238. Kinetic Mechanism ofEscherichiacoliAsparagine Synthetase B†. DOI:10.1021/bi981058h. PMID:9748330.
- Boehlein SK et al. (1997), J Biol Chem, 272, 12384-12392. Mutagenesis and Chemical Rescue Indicate Residues Involved in beta -Aspartyl-AMP Formation by Escherichia coli Asparagine Synthetase B. DOI:10.1074/jbc.272.19.12384. PMID:9139684.
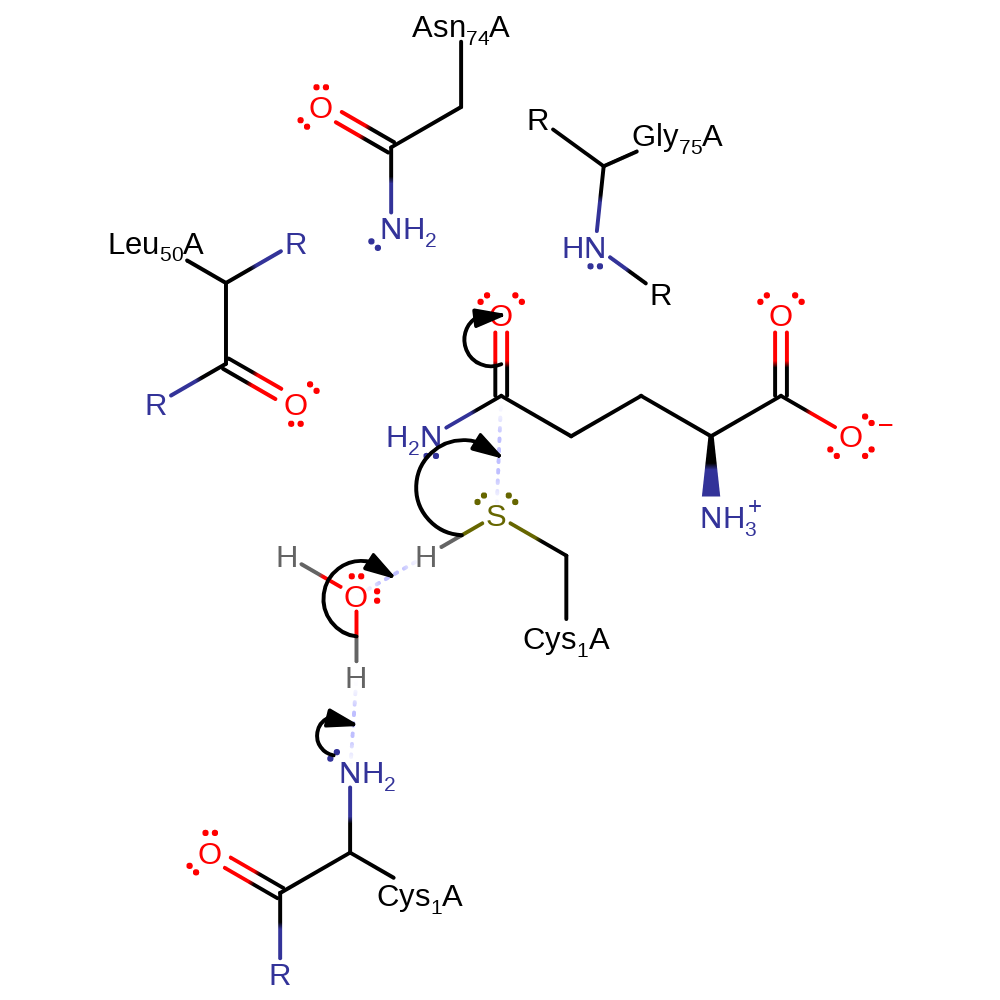
Step 1. This reaction takes place in the glutaminase domain. The N-terminus of Cys1 deprotonates water, which deprotonates the thiol group of Cys1, initiating a nucleophilic attack on the amide carbon in an addition reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Leu50A (main-C) | hydrogen bond acceptor, electrostatic stabiliser |
| Gly75A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Asn74A | hydrogen bond donor, electrostatic stabiliser |
| Ala1A (N-term) | proton acceptor |
| Ala1A | nucleophile, proton donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, proton relay, enzyme-substrate complex formation, overall reactant used, intermediate formation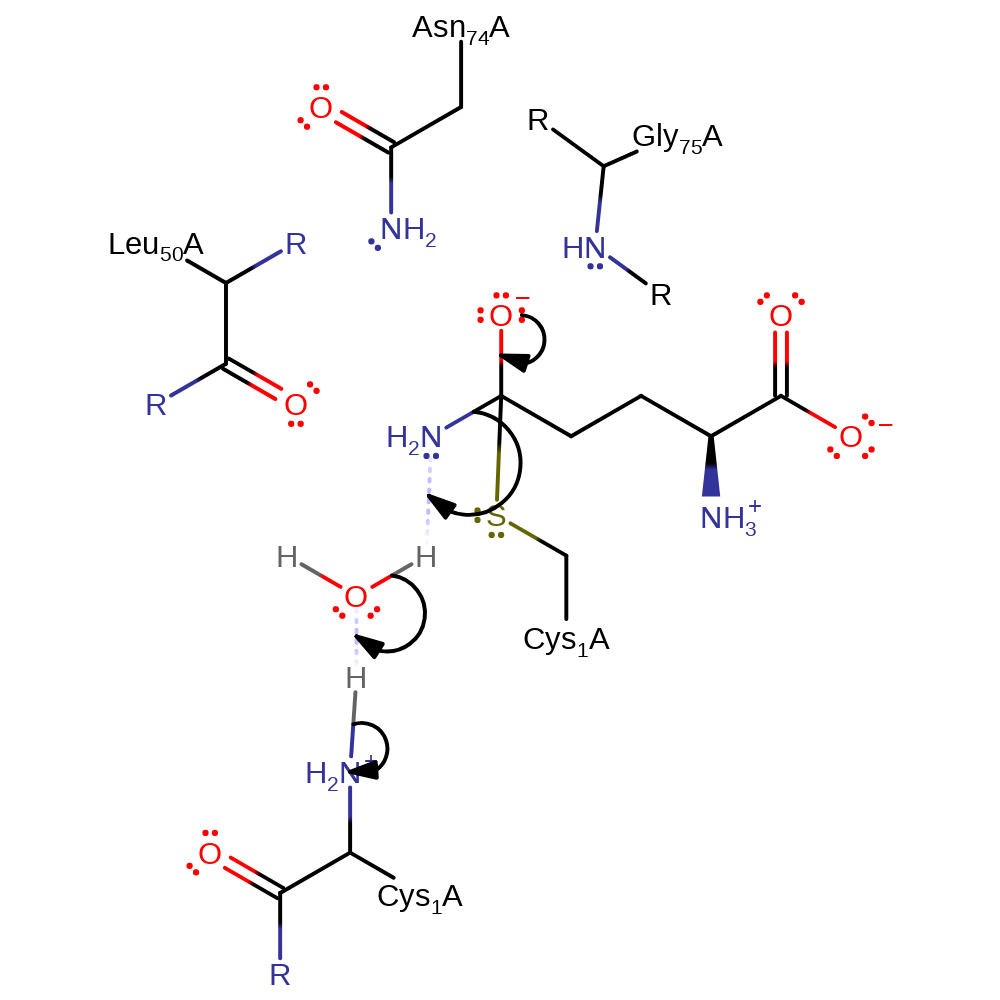
Step 2. This reaction takes place in the glutaminase domain. The oxyanion initiates an elimination that cleaves ammonia from the bound L-glutamine substrate. Ammonia deprotonates water, which deprotonates the N-terminus of Cys1. The ammonia product is then transferred to the second domain of the enzyme through an internal ion channel
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ala1A | covalently attached |
| Leu50A (main-C) | hydrogen bond acceptor, electrostatic stabiliser |
| Gly75A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Asn74A | hydrogen bond donor, electrostatic stabiliser |
| Ala1A (N-term) | proton donor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, proton relay, intermediate formation, enzyme-substrate complex cleavage, deamination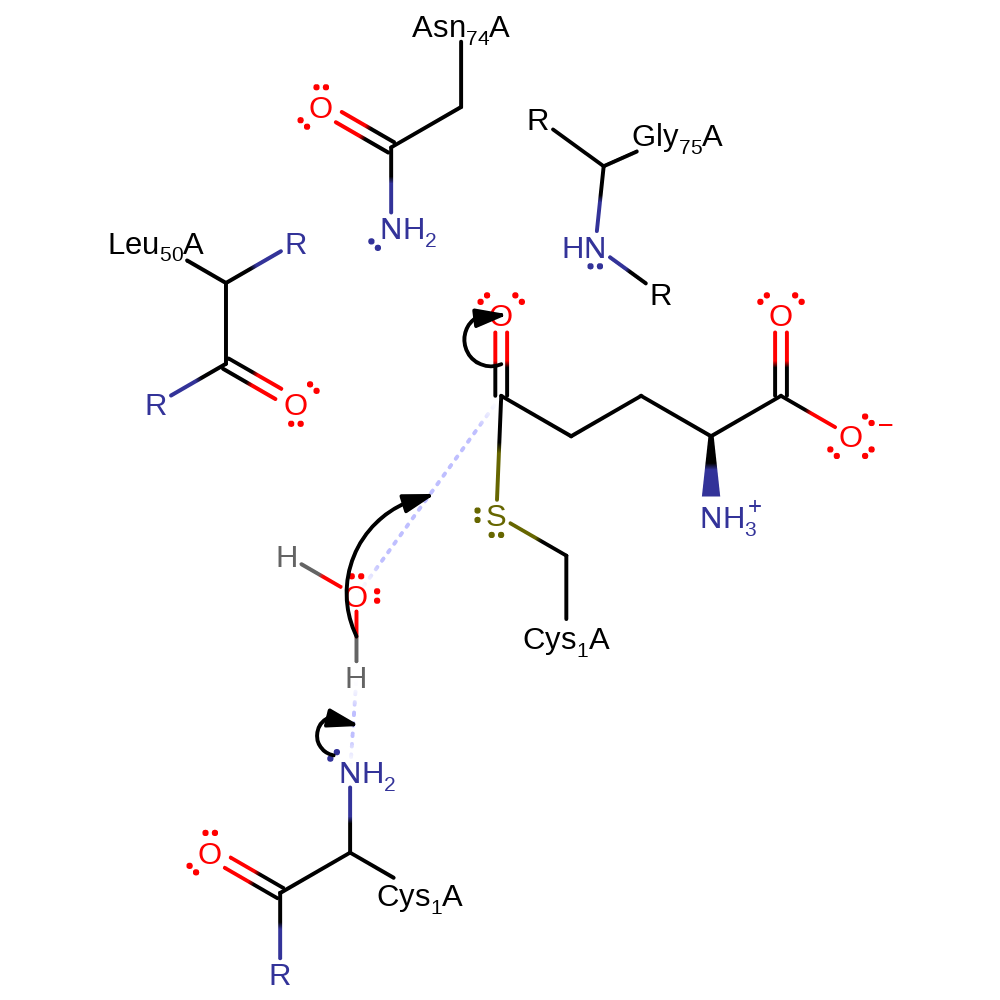
Step 3. This reaction occurs in the glutaminase domain. The N-terminus of Cys1 deprotonates water, which initiates a nucleophilic attack on the carbonyl carbon of the covalently bound intermediate in an addition reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ala1A | covalently attached, hydrogen bond acceptor |
| Leu50A (main-C) | hydrogen bond acceptor, electrostatic stabiliser |
| Gly75A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Asn74A | hydrogen bond donor, electrostatic stabiliser |
| Ala1A (N-term) | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, enzyme-substrate complex formation, overall reactant used, intermediate formation
Step 4. This reaction occurs in the glutaminase domain. The oxyanion initiates an elimination that cleaves the C-S bond, the thiolate of Cys1 deprotonates water, which deprotonates the N-terminus of Cys1.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ala1A | hydrogen bond acceptor |
| Leu50A (main-C) | hydrogen bond acceptor, electrostatic stabiliser |
| Gly75A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Asn74A | hydrogen bond donor, electrostatic stabiliser |
| Ala1A (N-term) | proton donor |
| Ala1A | nucleofuge, proton acceptor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, proton relay, intermediate collapse, overall product formed, enzyme-substrate complex cleavage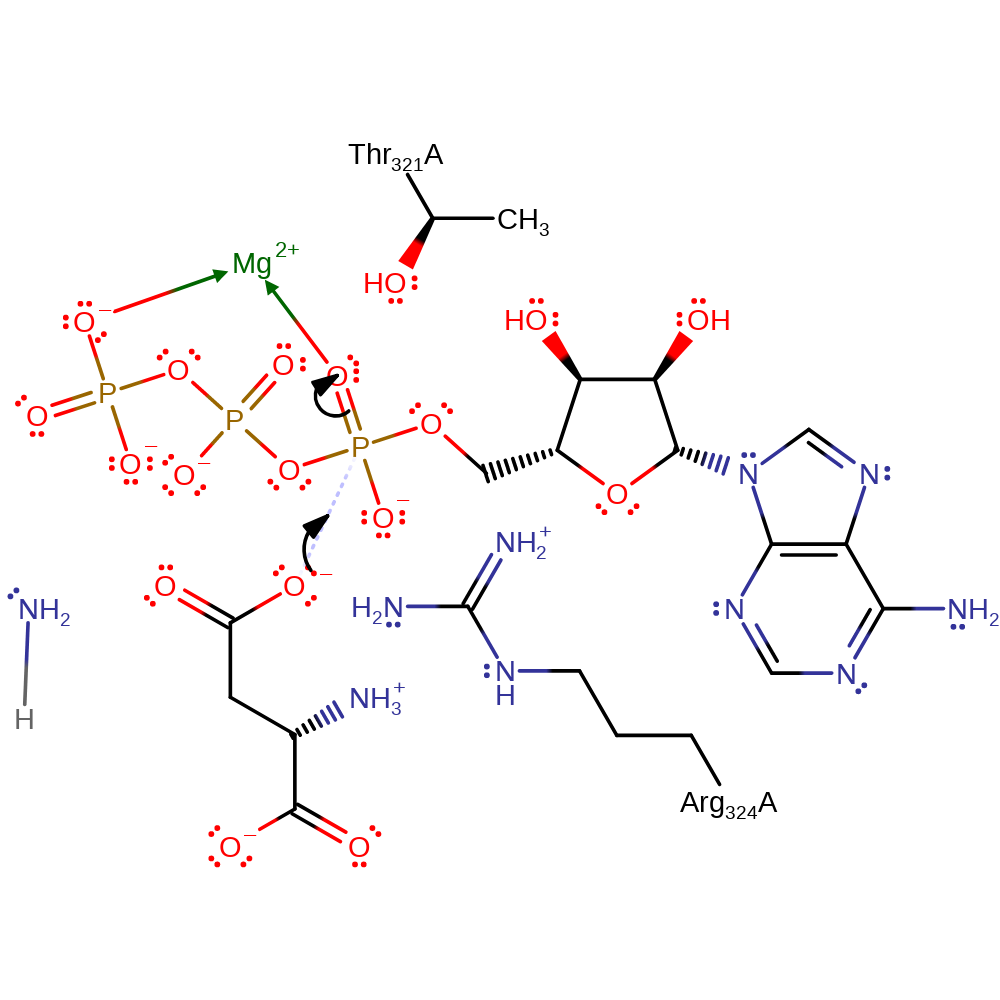
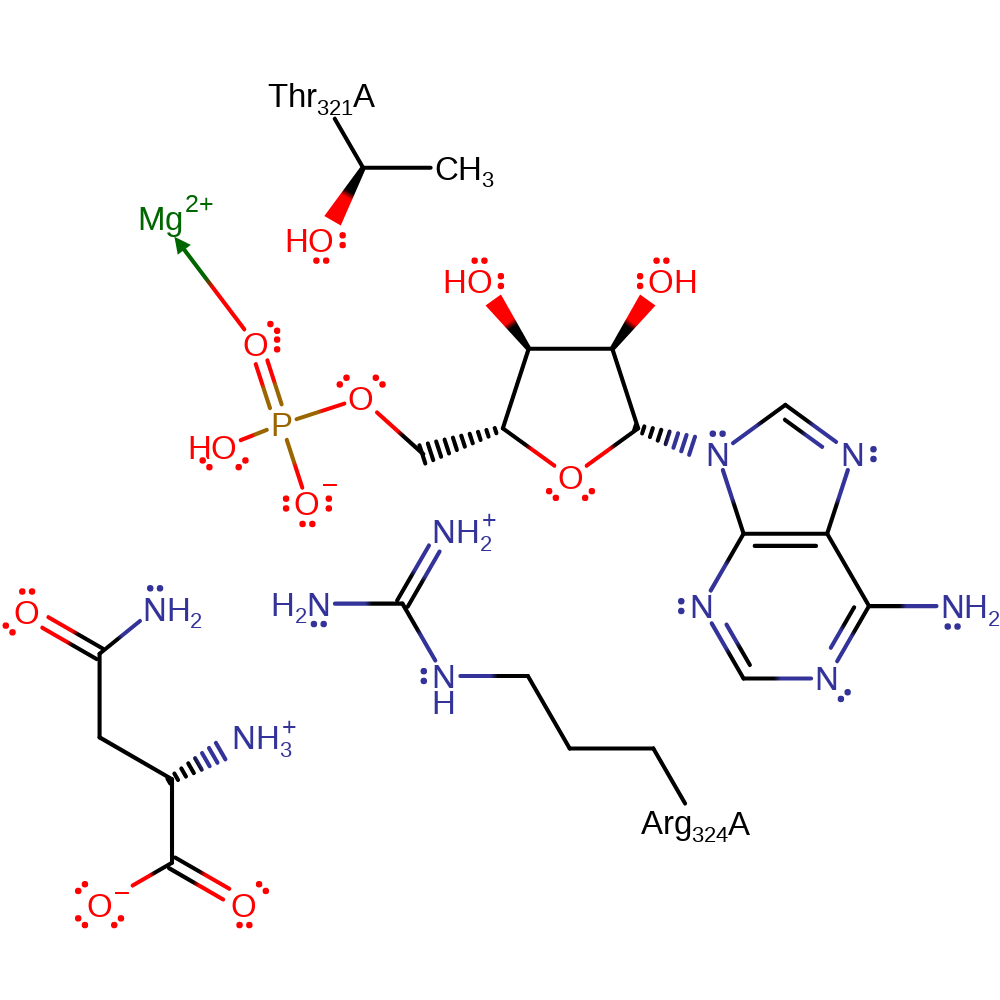
Step 5. This reaction occurs in the ligase domain of the enzyme. The aspartate substrate initiates a nucleophilic attack on the alpha phosphate of ATP in an addition reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg324A | hydrogen bond donor, electrostatic stabiliser |
| Thr321A | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
ingold: bimolecular nucleophilic addition, overall reactant used, intermediate formation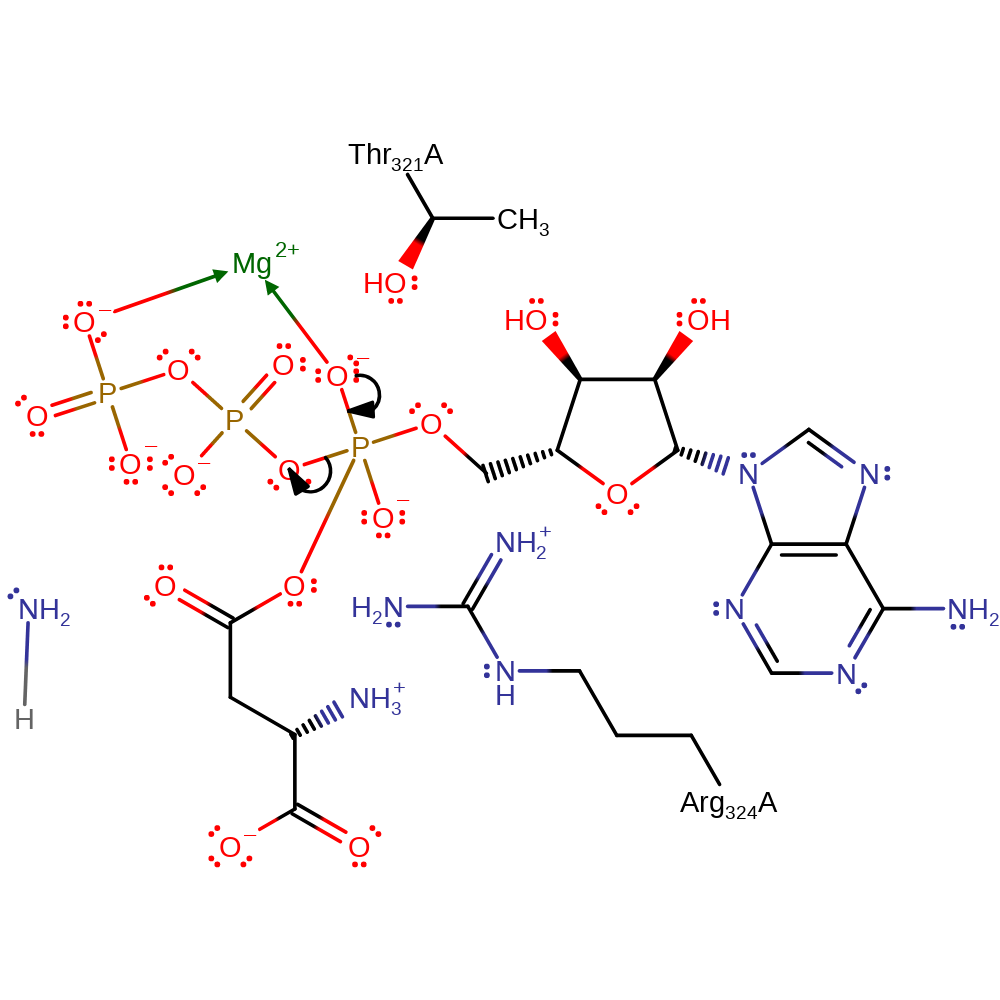
Step 6. This reaction occurs in the ligase domain of the enzyme. The pentavalent intermediate collapses to eliminate the diphosphate product and the beta-Asp-AMP intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg324A | hydrogen bond donor, electrostatic stabiliser |
| Thr321A | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
ingold: unimolecular elimination by the conjugate base, overall product formed, intermediate formation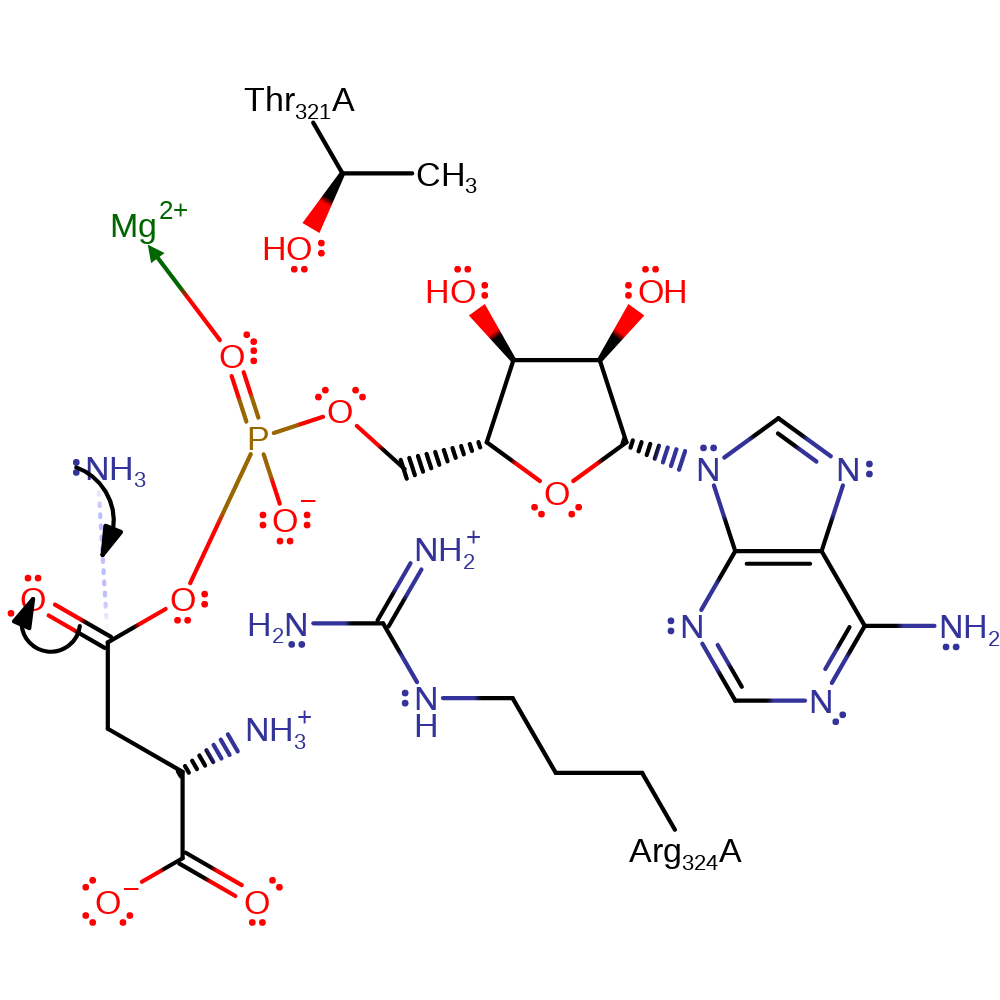
Step 7. This reaction occurs in the ligase domain of the enzyme. The ammonia nitrogen initiates a nucleophilic attack on the carbonyl carbon of the beta-Asp-AMP intermediate in an addition reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg324A | hydrogen bond donor, electrostatic stabiliser |
| Thr321A | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
ingold: bimolecular nucleophilic addition, intermediate formation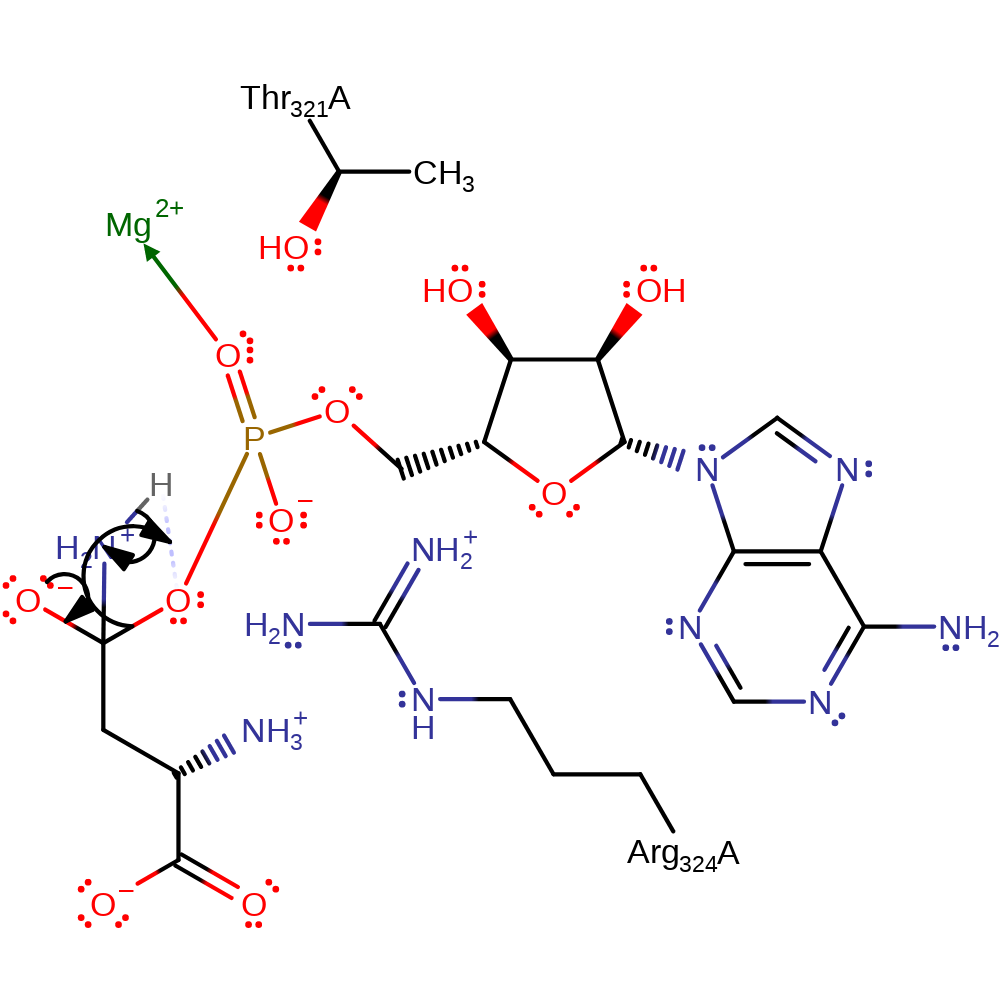
Step 8. This reaction occurs in the ligase domain of the enzyme. The oxyanion initiates an elimination that cleaves the C-O bond, the phosphate of AMP deprotonates the ammonium group, releasing the AMP and Asparagine products.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg324A | hydrogen bond donor, electrostatic stabiliser |
| Thr321A | hydrogen bond donor, electrostatic stabiliser |



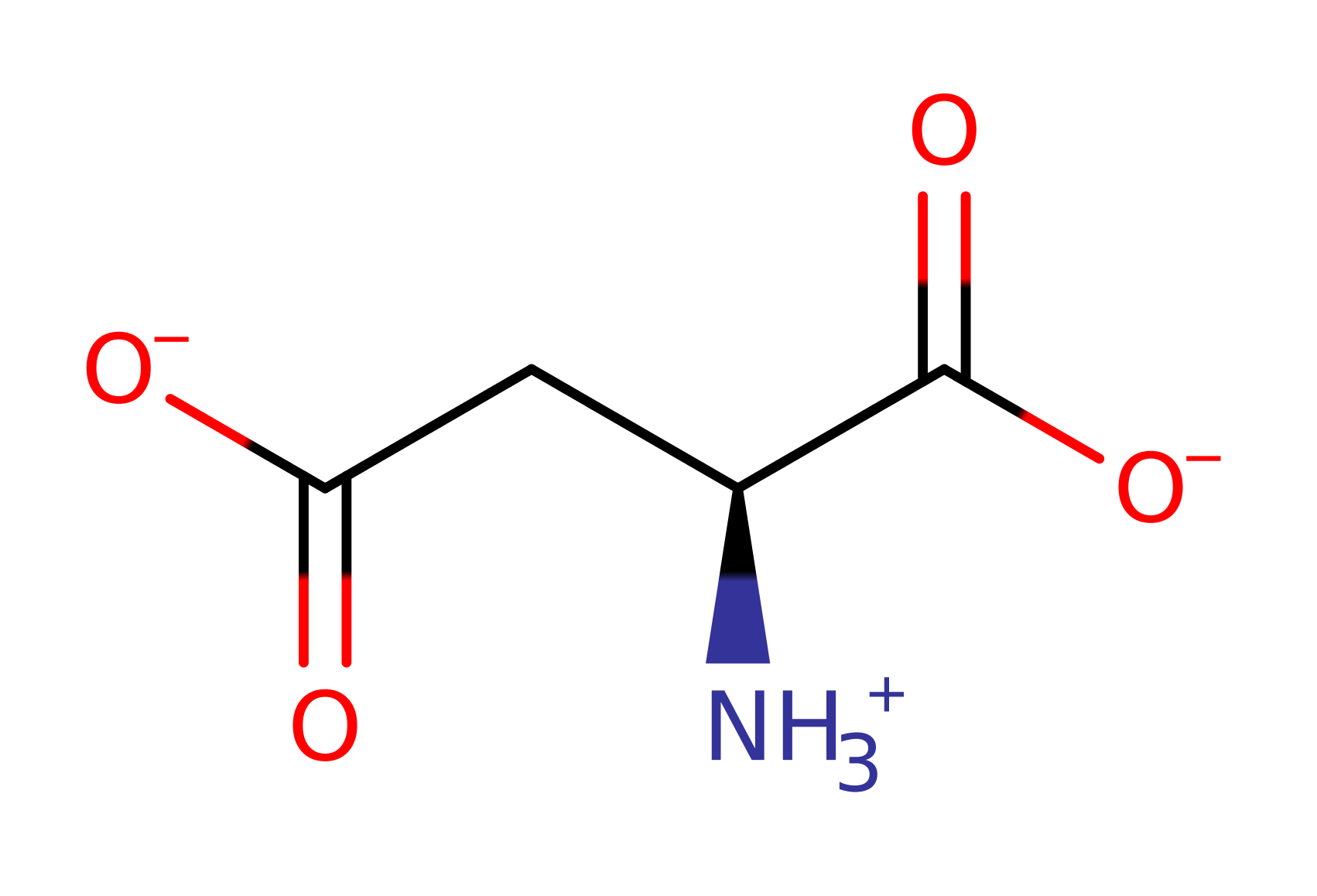





 Download:
Download: