Acetolactate synthase (biosynthetic)
Acetolactate synthase is a thiamin pyrophosphate-dependent enzyme that combines two molecules of pyruvate in a stereospecific condensation to yield 2-acetolactate with the release of carbon dioxide. Only the S enantiomer of 2-acetolactate is formed, which is then relayed into the biosynthesis of valine and leucine. Previously it was though that this protein also requires FAD for the protein to fold correctly, although the FAD is not involved in the reaction itself however recent studies (Lonhienne T et al.) have shown that it is involved in reaction by producing the radical that initiates this reaction. It exists in two distinct forms (biosynthetic and catabolic). This entry represents the biosynthetic form that is found in plants, fungi and bacteria.
Reference Protein and Structure
- Sequence
-
P07342
 (2.2.1.6)
(2.2.1.6)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Saccharomyces cerevisiae S288c (Baker's yeast)

- PDB
-
1n0h
- Crystal Structure of Yeast Acetohydroxyacid Synthase in Complex with a Sulfonylurea Herbicide, Chlorimuron Ethyl
(2.8 Å)



- Catalytic CATH Domains
-
3.40.50.970
 (see all for 1n0h)
(see all for 1n0h)
- Cofactors
- Thiamine(1+) diphosphate(3-) (1), Magnesium(2+) (1), Fadh2(2-) (1), Dioxygen (1) Metal MACiE
Enzyme Reaction (EC:2.2.1.6)
Enzyme Mechanism
Introduction
In this mechanism it is proposed that FAD and O2 are involved in an indirect one electron redox cycle. Studies have shown that the spatial configurations of O2 and FAD in the active site can allow electrons to be exchanged with the substrates and catalytic intermediates and also in the same paper, electron paramagnetic resonance evidence that a radical is produced during AHAS catalysis showing that the ligation of 2 pyruvates occurs via a radical mechanism (Lonhienne T et al.). The mechanism starts with FAD in the reduced form and ThDP is in its ylide form and concomitantly an electron is transferred from FAD to the donor pyruvate; and ThDP-ylide nucleophilically attacks the keto carbon; and also an electron is transferred from the carboxylic group of pyruvate to the cofactor oxygen. These transfers of electrons initiate the decarboxylation of pyruvate. The second "acceptor" pyruvate enters the active site and accepts an electron from the superoxide which initiates the carboligation of pyruvate to HE-ThDP and as result there is a series of electron transfers which result in the cleavage of ThDP from the now formed acetolactate. Also an electron is transferred back to FADH so that it can be regnerated.
Catalytic Residues Roles
| UniProt | PDB* (1n0h) | ||
| Phe201 | Phe201(191)A | Acts an electron relay from FADH to the keto oxygen of the donor pyruvate. | single electron relay, single electron acceptor, single electron donor |
| Gln202 | Gln202(192)A | Stabilises the formation of the superoxide | electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic addition, bimolecular homolytic elimination, electron relay, electron transfer, homolysis, intermediate formation, overall reactant used, radical formation, inferred reaction step, cofactor used, overall product formed, colligation, intermediate collapse, intermediate terminated, native state of cofactor regenerated, radical propagation, radical terminationReferences
- Lonhienne T et al. (2017), ChemistrySelect, 2, 11981-11988. High Resolution Crystal Structures of the Acetohydroxyacid Synthase-Pyruvate Complex Provide New Insights into Its Catalytic Mechanism. DOI:10.1002/slct.201702128.
- Liu Y et al. (2016), Appl Microbiol Biotechnol, 100, 8633-8649. Acetohydroxyacid synthases: evolution, structure, and function. DOI:10.1007/s00253-016-7809-9. PMID:27576495.
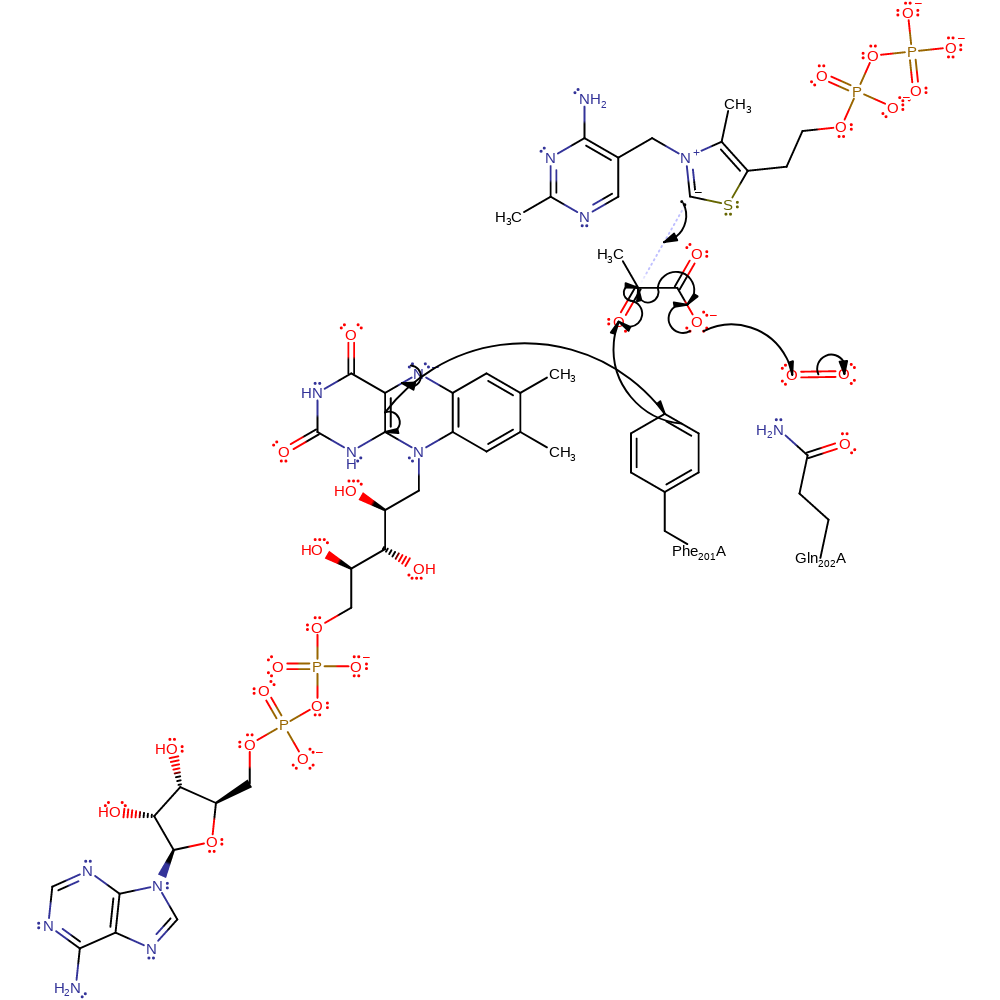
Step 1. An electron is transferred from FAD to the keto group of pyruvate and a second electron is transferred from the carboxylic group of pyruvate to oxygen. Also ThDP-ylide nucleophilically attack the electrophilic carbon atom of the keto group of pyruvate. These simultaneous reactions trigger the decarboxylation of pyruvate and the covalent binding of the aldehyde moiety of pyruvate to ThDP, forming HE-ThDP and CO2.The transfer of an electron from FAD to pyruvate can occur via Phe 201 which will act as an electron relay.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln202(192)A | electrostatic stabiliser |
| Phe201(191)A | single electron relay, single electron acceptor, single electron donor |
Chemical Components
ingold: bimolecular nucleophilic addition, ingold: bimolecular homolytic elimination, electron relay, electron transfer, homolysis, intermediate formation, overall reactant used, radical formation, inferred reaction step, cofactor used, overall product formed
Step 2. A second pyruvate enters the active site and accepts an electron from the radical oxygen cofactor. This initiates pyruvate to colligate to HE-ThDP which results in a series of electron transfers that result in the cleavage of the final product from ThDP and the transfer of an electron back to FADH(radical). Phe201 is though to act as an electron relay and transfer the single electron back from the substrate to FAD.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln202(192)A | electrostatic stabiliser |
| Phe201(191)A | single electron relay, single electron acceptor, single electron donor |
Chemical Components
ingold: bimolecular homolytic elimination, colligation, electron relay, electron transfer, homolysis, inferred reaction step, intermediate collapse, intermediate terminated, native state of cofactor regenerated, overall product formed, radical propagationCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
colligation, electron transfer, native state of cofactor regenerated, radical terminationIntroduction
Glu139 residue deprotonates the thiamine diphosphate cofactor at the N1 position. This initiates double bond rearrangement which results in the deprotonation of the N=CH-S group. This activates the cofactor towards electrophilic attack. The carbanion of thiamine diphosphate initiates a nucleophilic attack on the carbonyl carbon of pyruvate in an addition reaction. The conjugated double bond system of the cofactor undergoes rearrangement which results in the deprotonation of the glutamate residue. The covalently bound pyruvate undergoes decarboxylation. A second pyruvate molecule approaches the cofactor intermediate in a Si orientation and undergoes electrophilic addition at the carbonanion to give the S enantiomeric acetolacty l-ThDP. The acetolacty l-ThDP intermediate undergoes an intramolecular proton transfer with concurrent elimination of S-acetolactate. The TPP cofactor is regenerated by reprotonation of the C2 position.
Catalytic Residues Roles
| UniProt | PDB* (1n0h) | ||
| Lys251 | Lys251(241)A | Lys251B is directs the approach of the second pyruvate molecule by interacting with the carboxylic acid functionality. | steric locator |
| Glu139 | Glu139(129)A | Acts as a general acid/base in the activation of the thiamine diphosphate cofactor. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Gln202, Met582 | Gln202(192)A, Met582(572)B | The steric and electrostatic interactions between the intermediate and residues Met582and Gln202 holds the TPP cofactor in a high energy conformation which also contributes to enhanced reactivity. | hydrogen bond donor |
Chemical Components
proton transfer, cofactor used, intermediate formation, bimolecular nucleophilic addition, proton relay, unimolecular elimination by the conjugate base, overall product formed, decarboxylation, intermediate collapse, overall reactant used, elimination (not covered by the Ingold mechanisms), inferred reaction step, native state of cofactor regenerated, intermediate terminated, native state of enzyme regeneratedReferences
- Pang SS et al. (2004), J Biol Chem, 279, 2242-2253. The Crystal Structures of Klebsiella pneumoniae Acetolactate Synthase with Enzyme-bound Cofactor and with an Unusual Intermediate. DOI:10.1074/jbc.m304038200. PMID:14557277.
- Pang SS et al. (2003), J Biol Chem, 278, 7639-7644. Molecular Basis of Sulfonylurea Herbicide Inhibition of Acetohydroxyacid Synthase. DOI:10.1074/jbc.m211648200. PMID:12496246.
- Kern D et al. (1997), Science, 275, 67-70. How Thiamin Diphosphate Is Activated in Enzymes. DOI:10.1126/science.275.5296.67. PMID:8974393.
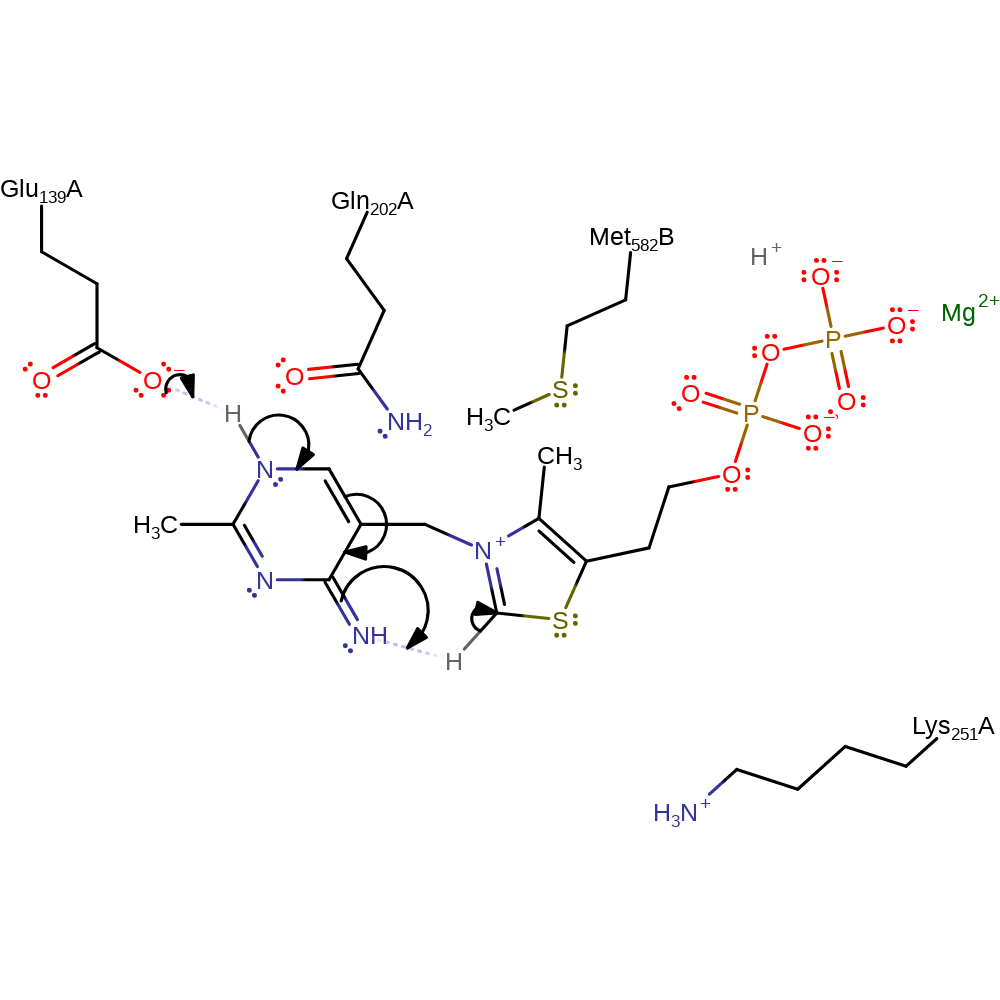
Step 1. A glutamate residue deprotonates the thiamine diphosphate cofactor at the N1 position. This initiates double bond rearrangement which results in the deprotonation of the N=CH-S group. This activates the cofactor towards electrophilic attack. Hydrogen bonding of the carboxylate group from an unspecified residue to the N1' of the aminopyrimidine ring increases the basic nature of the 4' position by stabilising the 4'-imino tautomeric form, allowing the group to deprotonate the C1 position of the thiamine ring [PMID:8974393]. Electron density in the crystal structure suggests that once the cofactor is deprotonated, a third, bridging ring, existing between the C2 and N4' positions contributes towards the possible resonance structures [PMID:14557277, PMID:8974393].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu139(129)A | hydrogen bond acceptor |
| Gln202(192)A | hydrogen bond donor |
| Met582(572)B | polar interaction, steric role |
| Glu139(129)A | proton acceptor |
Chemical Components
proton transfer, cofactor used, intermediate formation
Step 2. The carbanion of thiamine diphosphate initiates a nucleophilic attack on the carbonyl carbon of pyruvate in an addition reaction. The conjugated double bond system of the cofactor undergoes rearrangement which results in the deprotonation of the glutamate residue.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu139(129)A | hydrogen bond donor |
| Gln202(192)A | hydrogen bond donor |
| Met582(572)B | polar interaction, steric role |
| Glu139(129)A | proton donor |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, intermediate formation, proton relayCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu139(129)A | hydrogen bond acceptor |
| Gln202(192)A | hydrogen bond donor |
| Met582(572)B | polar interaction |
Chemical Components
ingold: unimolecular elimination by the conjugate base, overall product formed, decarboxylation, intermediate collapse, intermediate formation
Step 4. A second pyruvate molecule approaches the cofactor intermediate in a Si orientation and undergoes electrophilic addition at the carbonanion to give the S enantiomeric acetolacty l-ThDP.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu139(129)A | hydrogen bond acceptor |
| Gln202(192)A | hydrogen bond donor |
| Met582(572)B | polar interaction |
| Lys251(241)A | steric locator |
Chemical Components
ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used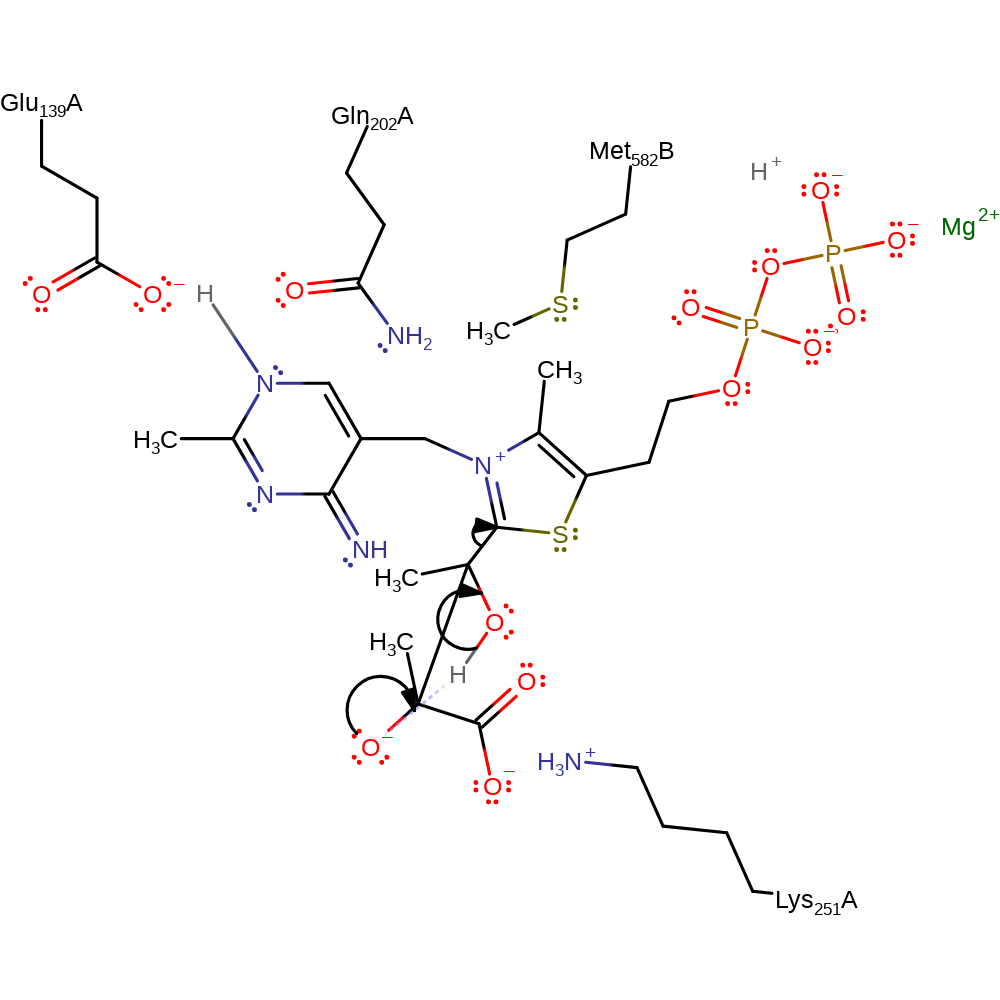
Step 5. The acetolacty l-ThDP intermediate undergoes an intramolecular proton transfer with concurrent elimination of S-acetolactate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln202(192)A | hydrogen bond donor |
| Met582(572)B | polar interaction |
Chemical Components
proton transfer, elimination (not covered by the Ingold mechanisms), intermediate collapse, overall product formed, inferred reaction step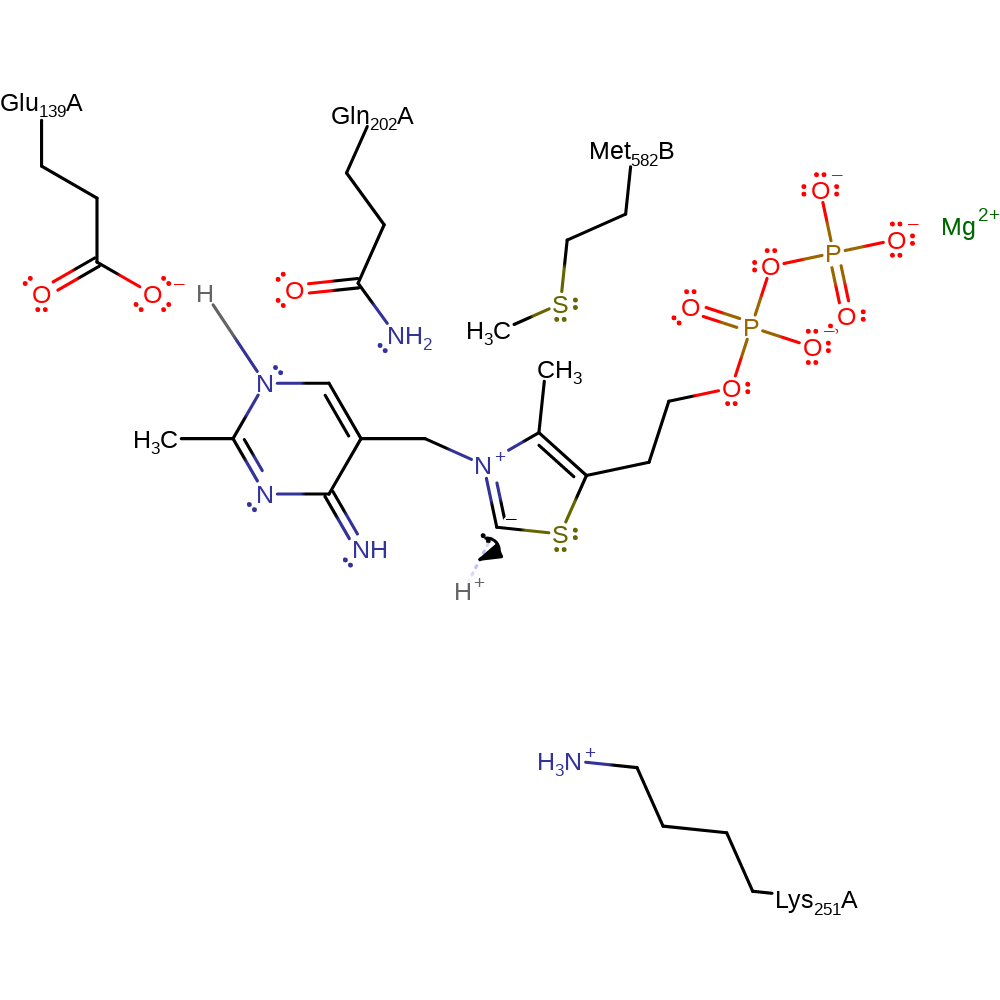
Step 6. The TPP cofactor is regenerated by reprotonation of the C2 position.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu139(129)A | hydrogen bond acceptor |
| Gln202(192)A | hydrogen bond donor |





 Download:
Download: 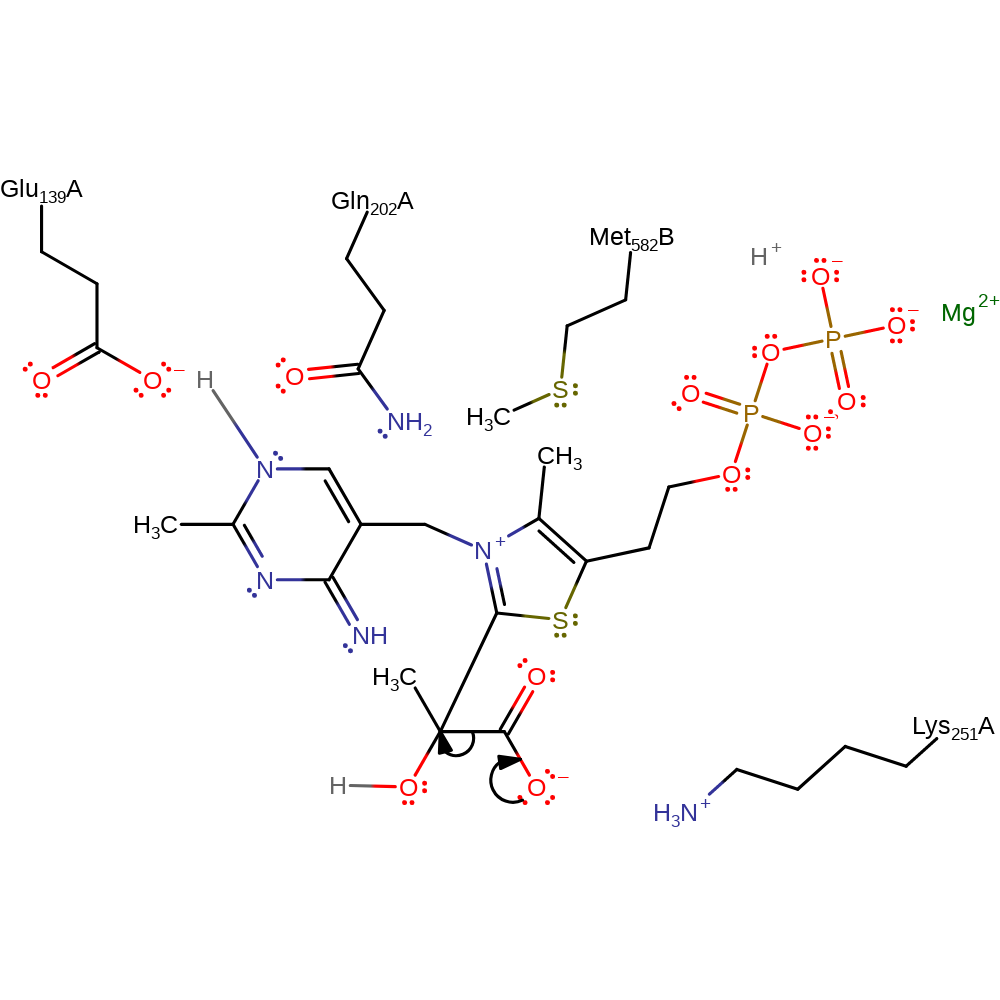
 Download:
Download: