Oligo-1,6-glucosidase
Oligo-1,6-glucosidase (dextrin 6-alpha-D-glucanohydrolase) hydrolyses the non-reducing terminal alpha-1,6-glucosidic bonds of isomaltosaccharides, panose, and alpha-limit dextrins, but fails to act on alpha,1,4-glucosidic bonds of maltosaccharides [PMID:8370659]. The anomeric configuration of the substrate is retained, thus the mechanism proceeds via a double displacement process [PMID:9193006, PMID:10331869]. In addition, this enzyme belongs to the family 13 of glycosyl hydrolases, also known as the alpha-amylase family.
Reference Protein and Structure
- Sequence
-
P21332
 (3.2.1.10)
(3.2.1.10)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Bacillus cereus (Bacteria)

- PDB
-
1uok
- CRYSTAL STRUCTURE OF B. CEREUS OLIGO-1,6-GLUCOSIDASE
(2.0 Å)



- Catalytic CATH Domains
-
3.20.20.80
 (see all for 1uok)
(see all for 1uok)
Enzyme Reaction (EC:3.2.1.10)
Enzyme Mechanism
Introduction
The mechanism here is shown utilising isomaltose. The alpha-retaining mechanism is a double displacement process, which proceeds in two steps. In the first step, Oligo-1,6-glucosidase cleaves an alpha(1-6) glycosidic bond in its substrate, dextrin or isomaltose, and forms a covalent beta(1-6)-linked glycosyl-enzyme intermediate [PMID:10331869]. In the second step, the resulting glycosyl enzyme is hydrolysed by a water molecule [PMID:11676021]. Two active site amino acids play distinct roles in catalysis. One is the acid/base Glu 255, which protonates the glycosidic oxygen of the scissile bond in the first step, and then deprotonates the attacking water molecule in the second step. The other is the nucleophile, Asp 229, which attacks the sugar, forming the covalent linkage within the intermediate [PMID:10331869, PMID:11676021].
Catalytic Residues Roles
| UniProt | PDB* (1uok) | ||
| Glu255 | Glu255A | Acts as a general acid/base. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, repulsive charge-charge interaction |
| His328 | His328A | Helps stabilise the reactive intermediates formed during the course of the reaction. | hydrogen bond donor, electrostatic stabiliser |
| Asp199 | Asp199A | Acts as the catalytic nucleophile. | covalently attached, nucleofuge, nucleophile, polar interaction |
| Asp98 | Asp98A | Acts to destabilise the enzyme-substrate complex ground state. | repulsive charge-charge interaction, electrostatic destabiliser, steric role, electrostatic stabiliser |
| Asp329 | Asp329A | Helps to stabilise the reactive intermediates and transition states formed during the course of the reaction. | transition state stabiliser |
Chemical Components
bimolecular nucleophilic substitution, proton transfer, enzyme-substrate complex formation, overall product formed, overall reactant used, intermediate formation, enzyme-substrate complex cleavage, intermediate terminated, native state of enzyme regeneratedReferences
- Uitdehaag JC et al. (1999), Nat Struct Biol, 6, 432-436. X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the alpha-amylase family. DOI:10.1038/8235. PMID:10331869.
- Hung VS et al. (2005), Appl Microbiol Biotechnol, 68, 757-765. alpha-Glucosidase from a strain of deep-sea Geobacillus: a potential enzyme for the biosynthesis of complex carbohydrates. DOI:10.1007/s00253-005-1977-3. PMID:15940457.
- Watanabe K et al. (2001), Biosci Biotechnol Biochem, 65, 2058-2064. Identification of Catalytic and Substrate-binding Site Residues in Bacillus cereus ATCC7064 Oligo-1,6-glucosidase. DOI:10.1271/bbb.65.2058. PMID:11676021.
- Watanabe K et al. (1997), J Mol Biol, 269, 142-153. The refined crystal structure of Bacillus cereus oligo-1,6-glucosidase at 2.0 å resolution: structural characterization of proline-substitution sites for protein thermostabilization. DOI:10.1006/jmbi.1997.1018. PMID:9193006.
- Kizaki H et al. (1993), J Biochem, 113, 646-649. Polypeptide folding of Bacillus cereus ATCC7064 oligo-1,6-glucosidase revealed by 3.0 A resolution X-ray analysis. PMID:8370659.
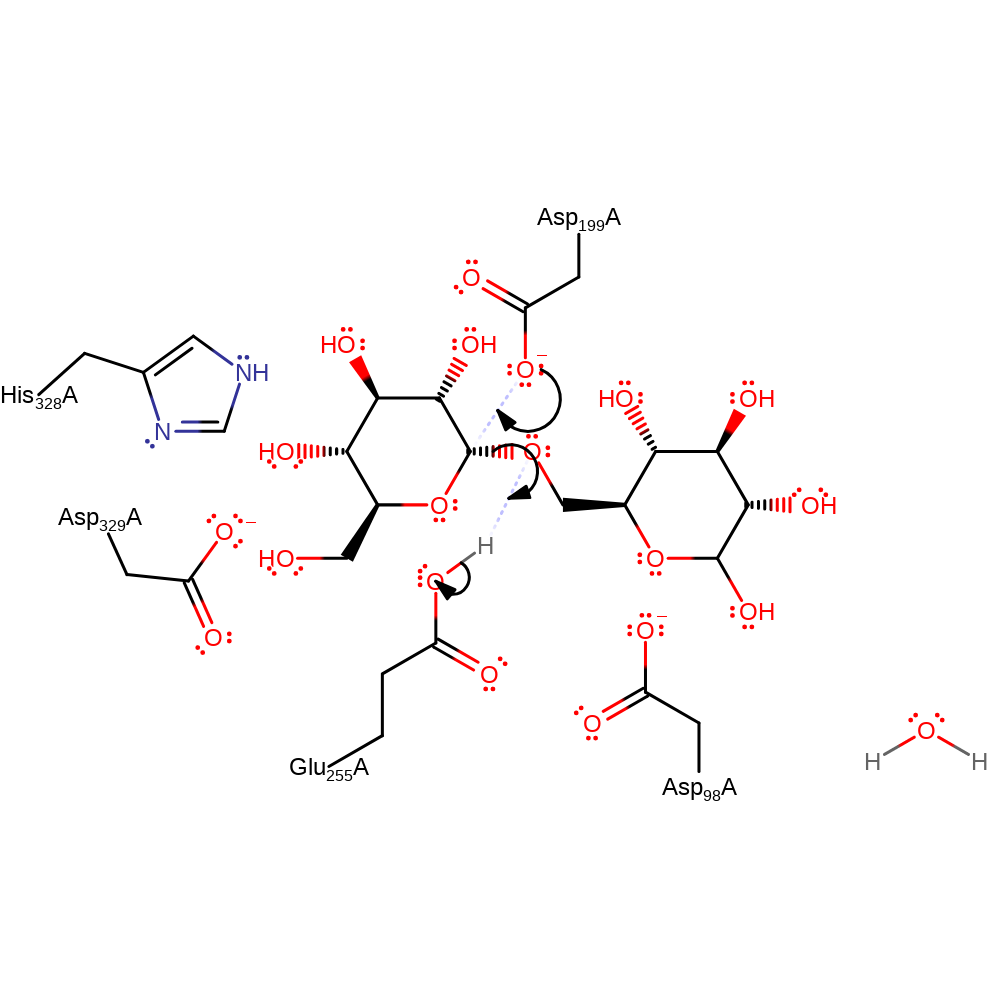
Step 1. Asp displaces the terminal sugar in a nucleophilic substitution reaction, forming a covalent bond between the enzyme and substrate. The mechanism proceeds through a high-energy oxocarbenium-like transition state, in which the reaction centre of the sugar is planar and positively charged [PMID:10331869].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp199A | polar interaction |
| His328A | electrostatic stabiliser, hydrogen bond donor |
| Asp98A | repulsive charge-charge interaction, electrostatic destabiliser, steric role, electrostatic stabiliser |
| Glu255A | hydrogen bond donor |
| Asp329A | transition state stabiliser |
| Asp199A | nucleophile |
| Glu255A | proton donor |
Chemical Components
ingold: bimolecular nucleophilic substitution, proton transfer, enzyme-substrate complex formation, overall product formed, overall reactant used, intermediate formation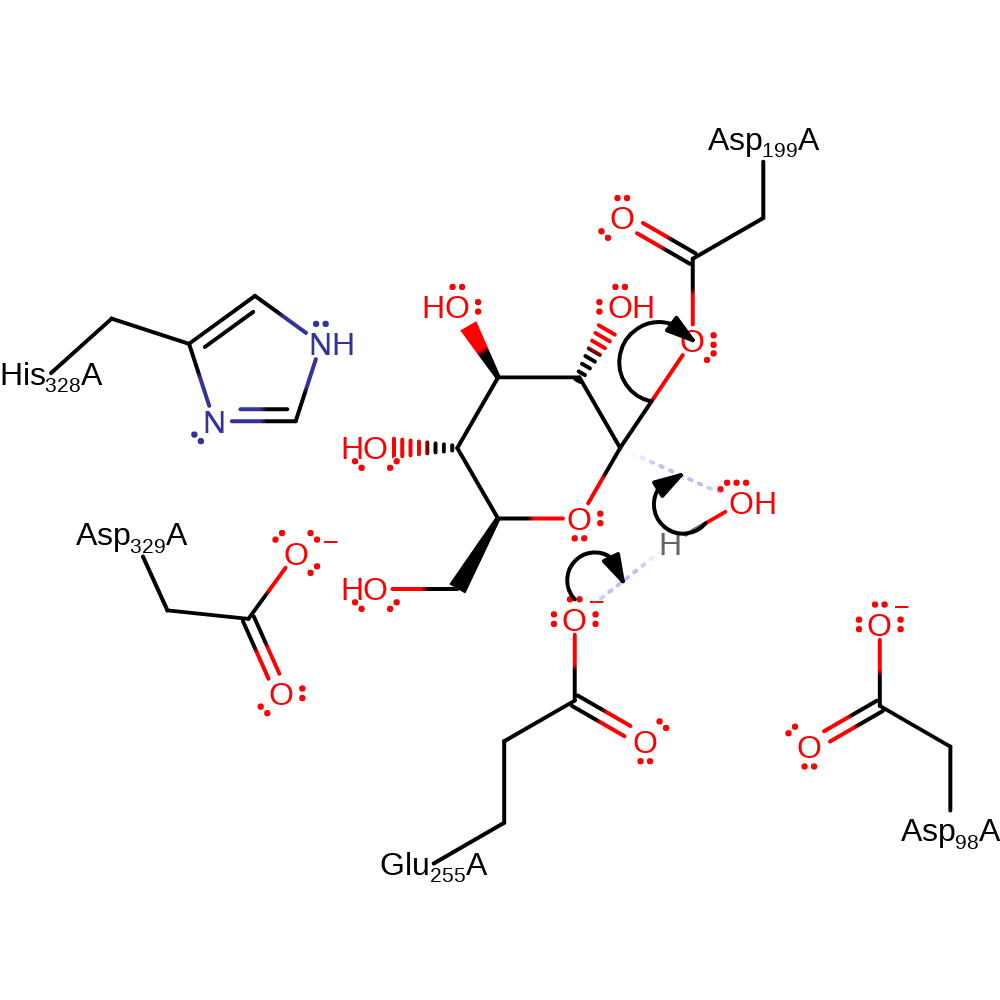
Step 2. Glu255 deprotonates the water, which initiates a nucleophilic attack on the covalently bound sugar, displacing the Asp199. The mechanism proceeds through a high-energy oxocarbenium-like transition state, in which the reaction centre of the sugar is planar and positively charged [PMID:10331869].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp199A | covalently attached |
| His328A | electrostatic stabiliser, hydrogen bond donor |
| Asp98A | repulsive charge-charge interaction, electrostatic destabiliser, steric role, electrostatic stabiliser |
| Glu255A | hydrogen bond acceptor, repulsive charge-charge interaction |
| Asp329A | transition state stabiliser |
| Glu255A | proton acceptor |
| Asp199A | nucleofuge |




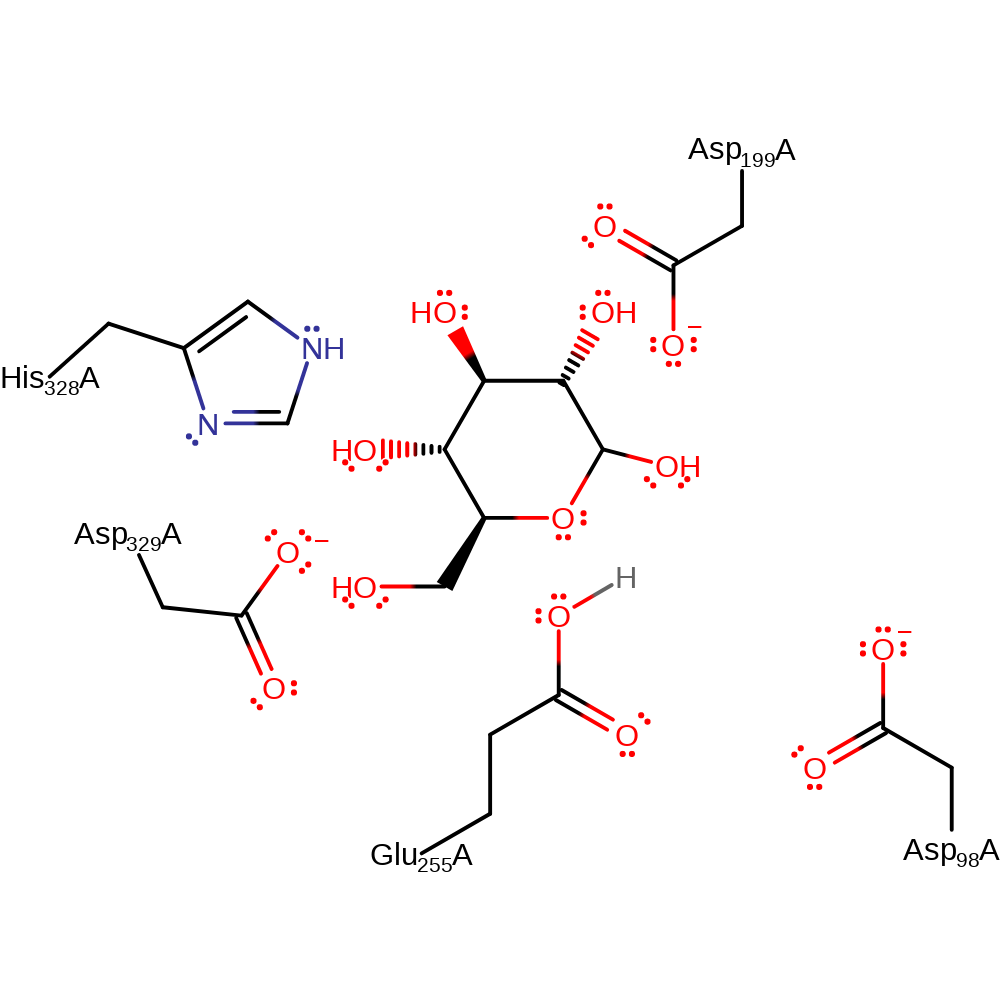 Download:
Download: