Methionine synthase
Methionine synthase (MetH) is a cobalamin (vitamin B12) and zinc dependent enzyme that catalyses the transfer of a methyl group from methyl-cobalamin to homocysteine, yielding enzyme-bound cob(I)alamin and methionine. The enzyme's cofactor is re-methylated using methyltetrahydrofolate. During the catalytic cycle, the highly reactive cob(I)alamin intermediate can be oxidised to produce an inactive cob(II)alamin enzyme; the enzyme is then reactivated via reductive methylation by the activation domain.
Reference Protein and Structure
- Sequence
-
P13009
 (2.1.1.13)
(2.1.1.13)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1bmt
- HOW A PROTEIN BINDS B12: A 3.O ANGSTROM X-RAY STRUCTURE OF THE B12-BINDING DOMAINS OF METHIONINE SYNTHASE
(3.0 Å)



- Catalytic CATH Domains
-
3.40.50.280
 (see all for 1bmt)
(see all for 1bmt)
- Cofactors
- Methylcobalamin (1) Metal MACiE
Enzyme Reaction (EC:2.1.1.13)
Enzyme Mechanism
Introduction
This enzyme is unique in E. coli in that it utilises methylcobalamin (rather than adenosylcobalamin) and catalysis involves a heterolytic (rather than homolytic) cleavage of the Co-C bond [PMID:8652590]. The Cob(I)alamin intermediate generated by this heterolytic cleavage is highly reactive and is occasionally oxidised to an inactive cob(II)alamin species. The enzyme must then re reactivated by reductive methylation to form methylcobalamin, which required S-adenosyl-methionine and an electron. In vitro, this oxidation occurs once every 100-2,000 turnovers, depending on the degree of anaerobiosis achieved during turnover. The physiological electron donor required for re-activation is believed to be flavodoxin and S-adenosyl-methionine is only required in catalytic amounts. [PMID:7992050, PMID:7712296]. It has been suggested that major domain rearrangements occur to allow the cobalamin reacts with its three different substrates [PMID:11731805]. Kinetic studies suggest that the methyltetrahydrofolate binds first to the enzyme, followed by homocysteine, which is then methylated by the cofactor, which is in turn re-methylated by the methyltetrahydrofolate [PMID:8652590].
Catalytic Residues Roles
| UniProt | PDB* (1bmt) | ||
| Asp757 | Asp757(107)A | Acts as a general acid/base and is part of a proton relay chain from bulk solvent to Ser810 and Hs759. Helps stabilise His759 as an anion in the enzyme's ground state. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, proton relay |
| His759 | His759(109)A | Acts as a general acid/base and is also the axial ligand to the cobalt ion of the cofactor. It is thought to be negatively charged in the enzyme's ground state, stabilised through a hydrogen bond network involving Asp757 and Ser810. | hydrogen bond acceptor, hydrogen bond donor, metal ligand, proton acceptor, proton donor, increase electrophilicity, increase nucleophilicity |
| Ser810 | Ser810(160)A | Acts as a general acid/base. Part of a proton relay chain from bulk solvent to Asp757 (then to His759). | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, proton relay |
Chemical Components
bimolecular nucleophilic substitution, proton transfer, charge delocalisation, cofactor used, intermediate formation, overall reactant used, overall product formed, proton relay, elimination (not covered by the Ingold mechanisms), intermediate terminated, native state of enzyme regenerated, native state of cofactor regeneratedReferences
- Drennan CL et al. (1994), Science, 266, 1669-1674. How a protein binds B12: A 3.0 A X-ray structure of B12-binding domains of methionine synthase. DOI:10.1126/science.7992050. PMID:7992050.
- Kumar N et al. (2013), J Phys Chem B, 117, 16044-16057. Mechanistic insights for formation of an organometallic Co-C bond in the methyl transfer reaction catalyzed by methionine synthase. DOI:10.1021/jp4093145. PMID:24164324.
- Bandarian V et al. (2002), Nat Struct Biol, 9, 53-56. Domain alternation switches B12-dependent methionine synthase to the activation conformation. DOI:10.1038/nsb738. PMID:11731805.
- Smith AE et al. (2000), Biochemistry, 39, 13880-13890. Protonation State of Methyltetrahydrofolate in a Binary Complex with Cobalamin-Dependent Methionine Synthase†. DOI:10.1021/bi001431x. PMID:11076529.
- Jarrett JT et al. (1996), Biochemistry, 35, 2464-2475. Mutations in the B12-Binding Region of Methionine Synthase: How the Protein Controls Methylcobalamin Reactivity†. DOI:10.1021/bi952389m. PMID:8652590.
- Drennan CL et al. (1994), Curr Opin Struct Biol, 4, 919-929. Cobalamin-dependent methionine synthase: the structure of a methylcobalamin-binding fragment and implications for other B12-dependent enzymes. DOI:10.1016/0959-440x(94)90275-5. PMID:7712296.
- Banerjee RV et al. (1990), FASEB J, 4, 1450-1459. Cobalamin-Dependent Methionine Synthase. DOI:10.1002/9781119951438.eibc0600. PMID:2407589.
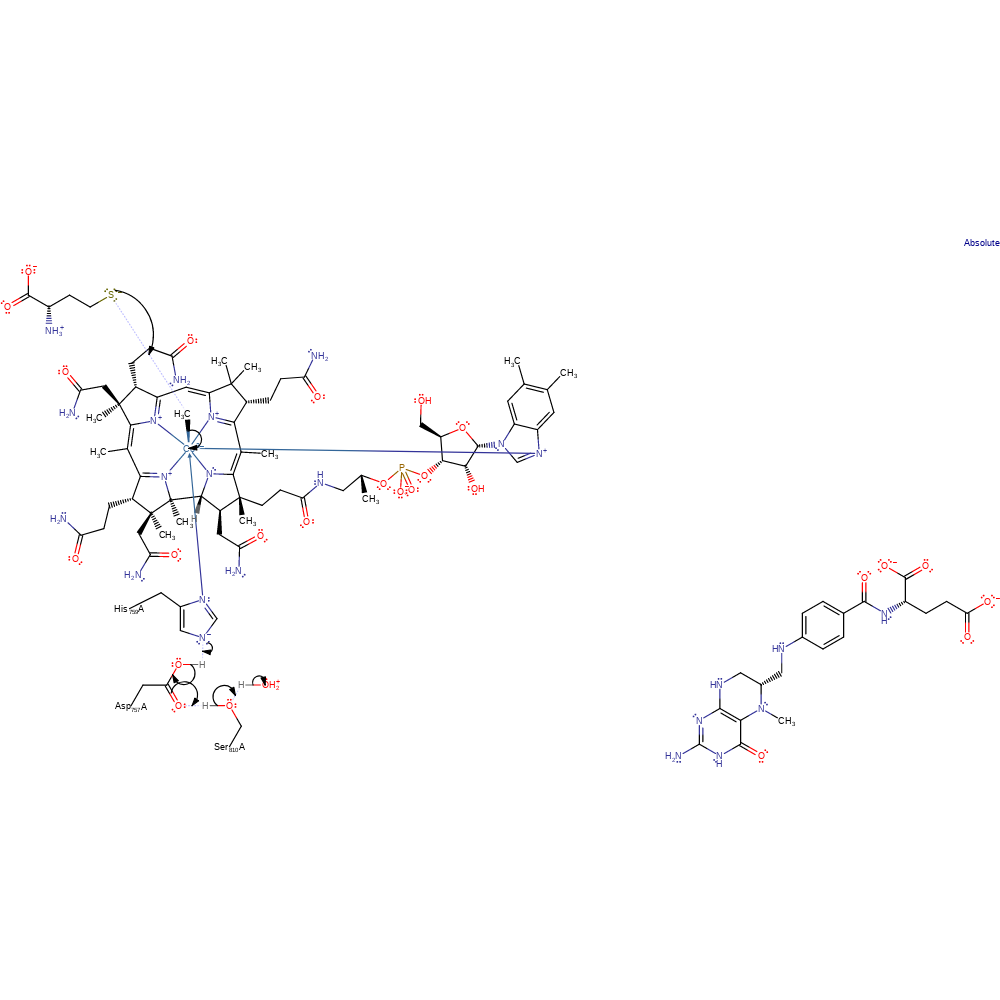
Step 1. The sulfur of homocysteine (assumed to bind in the deprotonated form) initiates a nulceophilic attack on the methyl of methylcobalamin, forming Co(I)alamin and the methionine product in a substitution reaction. A proton relay between His759 and bulk solvent modulates the reactivity of the cobalamin cofactor. In the methylated form of the enzyme, His759 and Asp757 share a single proton with one negative charge delocalised over two residues. Partial deprotonation of His757 greatly increases the bascicity of the ligand, stabilising the methylated form of the cofactor. [PMID:7992050]
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His759(109)A | metal ligand, increase electrophilicity, hydrogen bond acceptor |
| Asp757(107)A | hydrogen bond acceptor, hydrogen bond donor, proton relay |
| Ser810(160)A | hydrogen bond acceptor, hydrogen bond donor, proton relay |
| Asp757(107)A | proton acceptor, proton donor |
| His759(109)A | proton acceptor |
| Ser810(160)A | proton acceptor, proton donor |
Chemical Components
ingold: bimolecular nucleophilic substitution, proton transfer, charge delocalisation, cofactor used, intermediate formation, overall reactant used, overall product formed, proton relay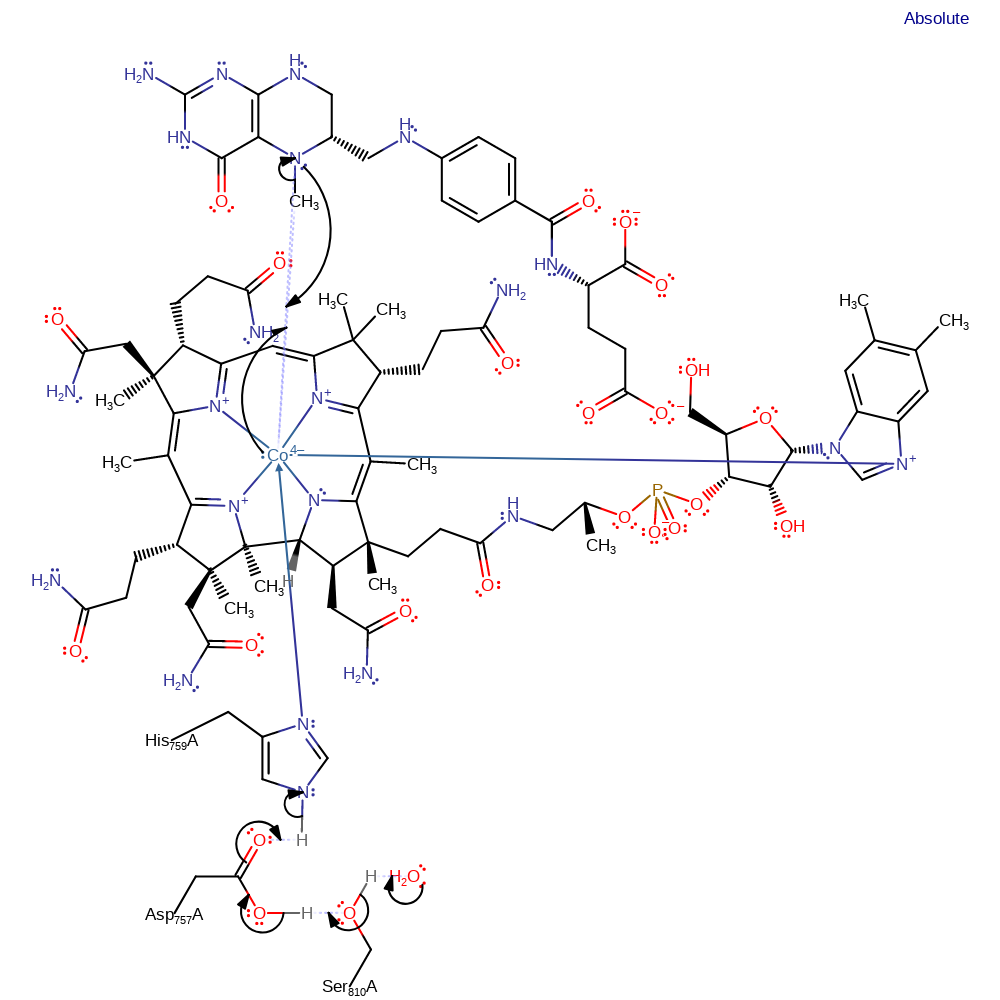
Step 2. The cob(I)alamin initiates a nucleophilic attack on the methyl group of methyltetrahydrofolate, proceeding through a three centre two electron bond, to form the methylcobalamin and covalently bound tetrahydrofolate. It is assumed that this reaction proceeds via a three center, two electron bond between the N, Co and C of the methyl group [PMID:11076529].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His759(109)A | metal ligand, increase nucleophilicity, hydrogen bond donor |
| Asp757(107)A | hydrogen bond acceptor, hydrogen bond donor, proton relay |
| Ser810(160)A | hydrogen bond acceptor, hydrogen bond donor, proton relay, proton donor |
| Asp757(107)A | proton acceptor, proton donor |
| Ser810(160)A | proton acceptor |
| His759(109)A | proton donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, charge delocalisation, intermediate formation, overall reactant used, proton relay
Step 3. The tetrahydrofolate product dissociated from the cobalamin cofactor with protonation (assumed to be from the bulk solvent). It is not clear when the protonation occurs, it is thought that protonation of the leaving group is esential for the cleavage of the Co-N bond. It is assumed that this proton comes from the bulk solvent. [PMID:11076529]
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His759(109)A | metal ligand, increase nucleophilicity |
| Asp757(107)A | hydrogen bond acceptor, hydrogen bond donor |
| Ser810(160)A | hydrogen bond acceptor, hydrogen bond donor |




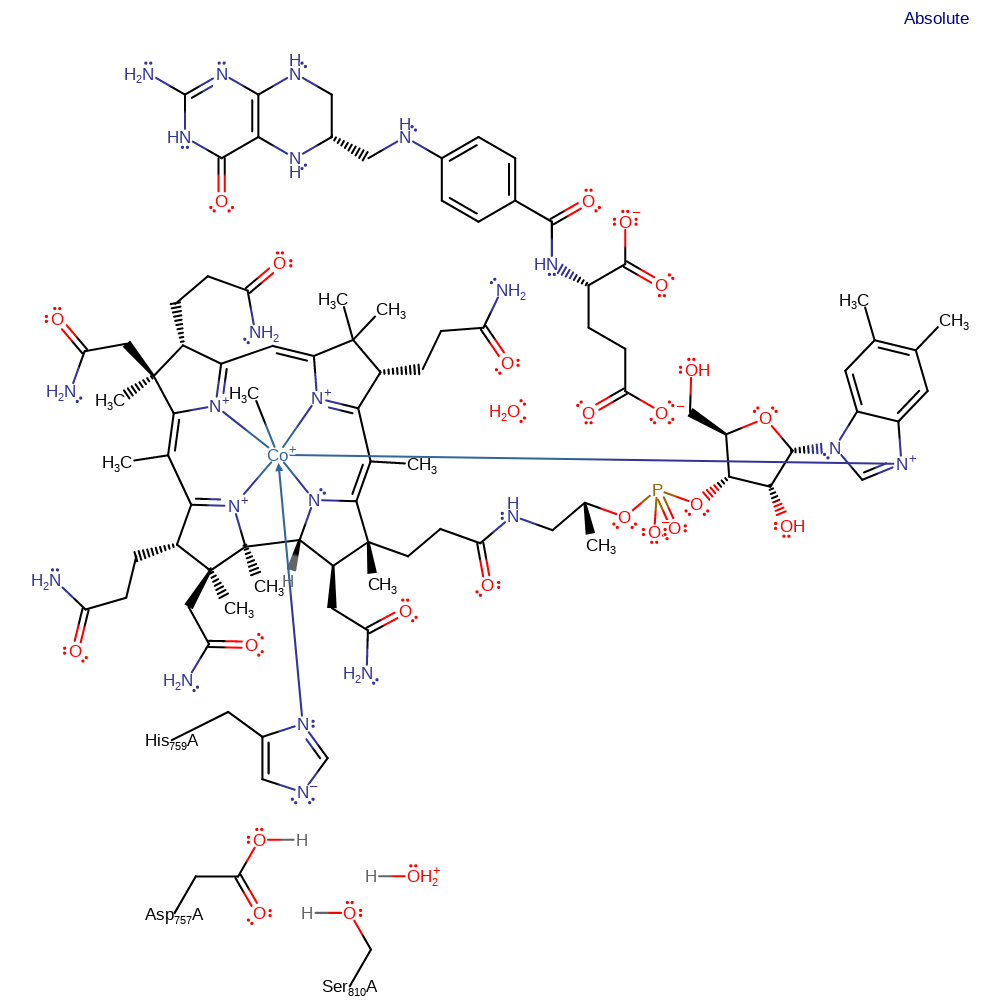 Download:
Download: