N4-(beta-N-acetylglucosaminyl)-L-asparaginase
Aspartylglucosylaminidase hydrolyses the bonds connecting N-linked oligosaccharides to free (not peptide bonded) asparagine residues. This is an important lysosomal function.
Aspartylglucosylaminidase is synthesised as a precursor protein which is activated by proteolytic cleavage between Asp 182 and Thr 183 to generate the required N-terminal threonine residue. The cleavage generates the alpha and beta subunits of the (alpha)2(beta)2 heterotetramer. The cleavage event is thought to be autocatalytic, and many of the residues involved in the normal glucosylaminidase are believed to be involved in the autocatalytic reaction. The Thr 183 side chain, for example, functions as a nucleophile in both [PMID:14616088].
Reference Protein and Structure
- Sequence
-
P20933
 (3.5.1.26)
(3.5.1.26)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Homo sapiens (Human)

- PDB
-
1apy
- HUMAN ASPARTYLGLUCOSAMINIDASE
(2.0 Å)



- Catalytic CATH Domains
-
3.60.20.30
 (see all for 1apy)
(see all for 1apy)
Enzyme Reaction (EC:3.5.1.26)
Enzyme Mechanism
Introduction
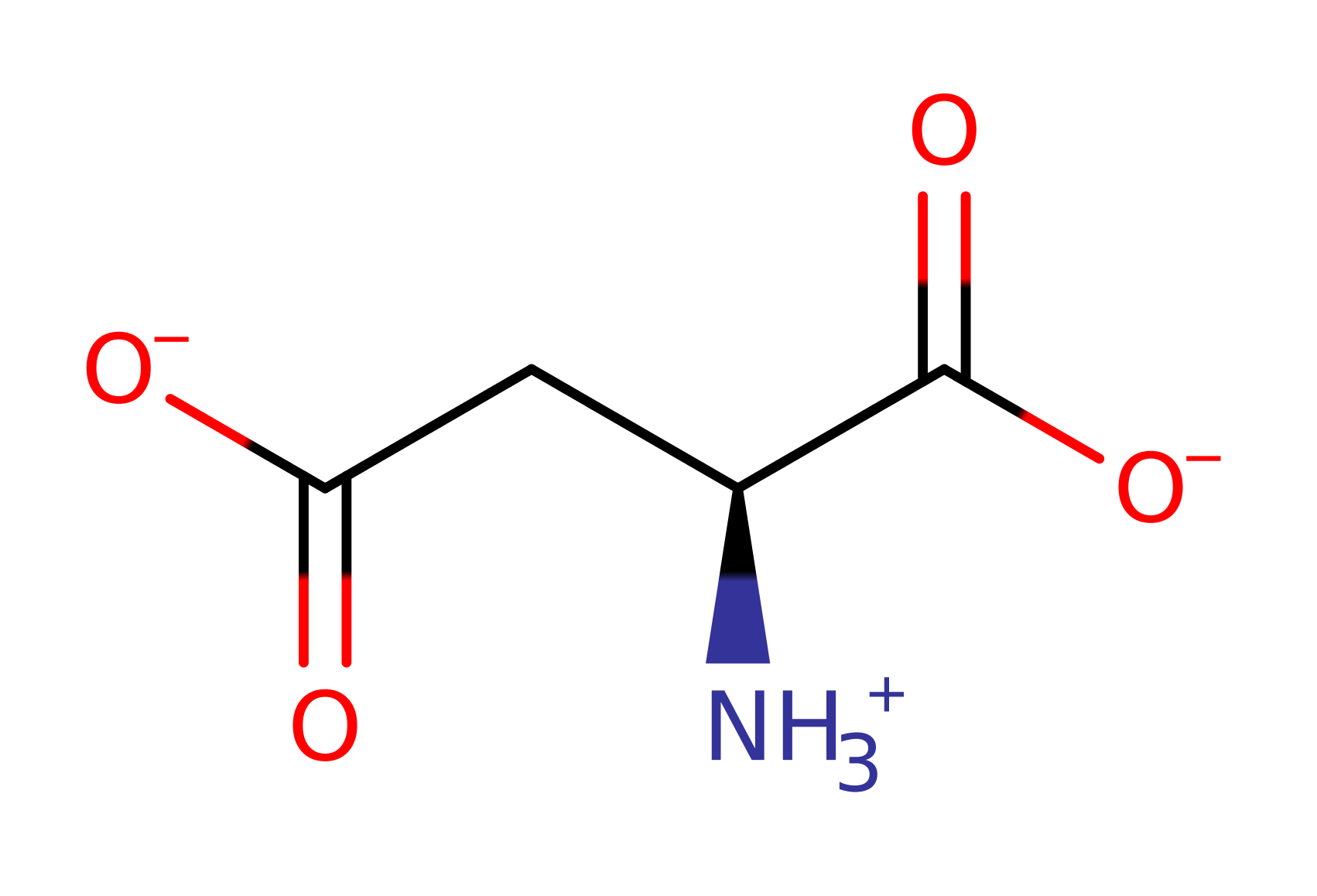
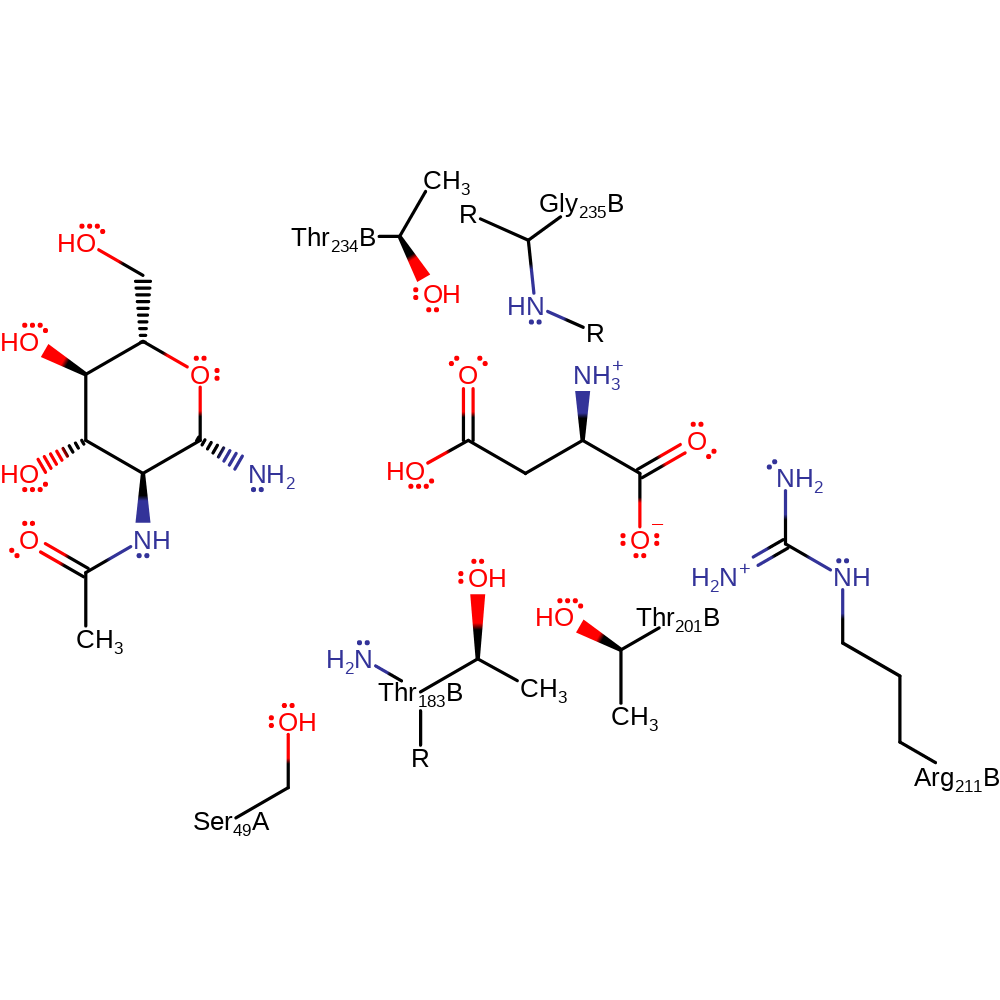
The sugar-asparagine substrate binds in the active site pocket, and is attacked by the hydroxyl group of an N-terminal threonine, Thr 206. The N-terminal amino group acts as a base in extracting the threonine's O-gamma proton allowing it to attack the substrate carbonyl group. The catalytic properties of Thr 206 are modified by Thr 224, which interacts with the Thr 206 O-gamma, and by Ser 72, which interacts with the Thr 206 alpha amino group. The tetrahedral intermediate that results from the nucleophilic attack is stabilised by an oxyanion hole composed of the side chain of Thr 257 and the backbone NH of Gly 258. Collapse of the tetrahedral intermediate with protonation of the departing amino group by the alpha amino group of Thr 206 generates an acyl-enzyme intermediate. This is then hydrolysed by a water molecule that is deprotonated by the alpha amino group of Thr 206.
Catalytic Residues Roles
| UniProt | PDB* (1apy) | ||
| Ser72 | Ser49A | Forms a hydrogen bond with the alpha amino group of Thr 206 and modifies the catalytic properties of Thr 206. | activator, hydrogen bond acceptor |
| Thr206 | Thr183(1)B | Side chain hydroxyl acts as a nucleophile to attack the amide carbonyl of the N-glycosidic bond. Alpha amino group acts as a base to deprotonate the side chain hydroxyl to allow the nucleophilic attack. It also protonates the departing amine leaving group of the sugar, and later deprotonates a water molecule during hydrolysis of the acyl enzyme intermediate. | covalently attached, hydrogen bond acceptor, hydrogen bond donor, nucleophile, nucleofuge, proton donor, proton acceptor |
| Thr224 | Thr201(19)B | Side chain OH interacts with the O-gamma of Thr 206, modifying the catalytic properties of Thr 206. | activator, hydrogen bond donor |
| Arg234 | Arg211(29)B | Binds the terminal carboxylate of the substrate, holding it in position and helping to stabilise the reactive intermediates formed. | hydrogen bond acceptor, electrostatic stabiliser |
| Gly258 (main-N) | Gly235(53)B (main-N) | Backbone NH forms part of the oxyanion hole that stabilises the tetrahedral intermediate. | hydrogen bond donor, electrostatic stabiliser |
| Thr257 | Thr234(52)B | Side chain OH forms part of the oxyanion hole that stabilises the tetrahedral intermediate. | electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation, intermediate formation, unimolecular elimination by the conjugate base, enzyme-substrate complex cleavage, intermediate collapse, overall product formed, intermediate terminated, native state of enzyme regeneratedReferences
- Tikkanen R et al. (1996), EMBO J, 15, 2954-2960. Functional analyses of active site residues of human lysosomal aspartylglucosaminidase: implications for catalytic mechanism and autocatalytic activation. PMID:8670796.
- Saarela J et al. (2004), Biochem J, 378, 363-371. Autoproteolytic activation of human aspartylglucosaminidase. DOI:10.1042/bj20031496. PMID:14616088.
- Peräkylä M et al. (1997), J Am Chem Soc, 119, 1189-1196. A Simulation of the Catalytic Mechanism of Aspartylglucosaminidase Usingab InitioQuantum Mechanics and Molecular Dynamics. DOI:10.1021/ja9628967.
- Oinonen C et al. (1995), Nat Struct Biol, 2, 1102-1108. Three-dimensional structure of human lysosomal aspartylglucosaminidase. DOI:10.1038/nsb1295-1102. PMID:8846222.
- Fisher KJ et al. (1993), FEBS Lett, 323, 271-275. Post-translational processing and Thr-206 are required for glycosylasparaginase activity. DOI:10.1016/0014-5793(93)81355-4. PMID:8500622.
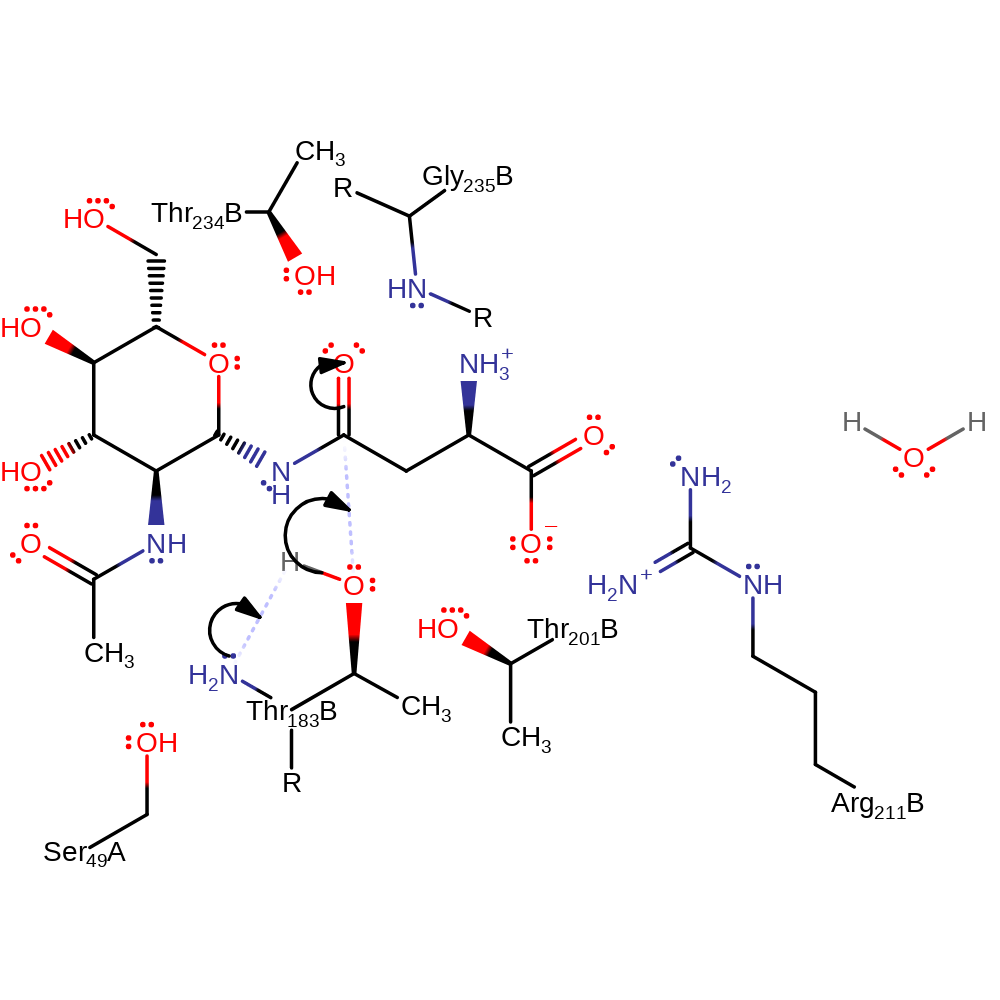
Step 1. The N-terminus of Thr206 deprotonates the alcohol group of Thr206, which attacks the carbonyl of the substrate in a nucleophilic addition.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly235(53)B (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Arg211(29)B | hydrogen bond acceptor, electrostatic stabiliser |
| Thr183(1)B | hydrogen bond donor |
| Ser49A | hydrogen bond acceptor, activator |
| Thr201(19)B | hydrogen bond donor, activator |
| Thr234(52)B | electrostatic stabiliser |
| Thr183(1)B | nucleophile |
| Thr183(1)B (N-term) | proton acceptor |
| Thr183(1)B | proton donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation, intermediate formation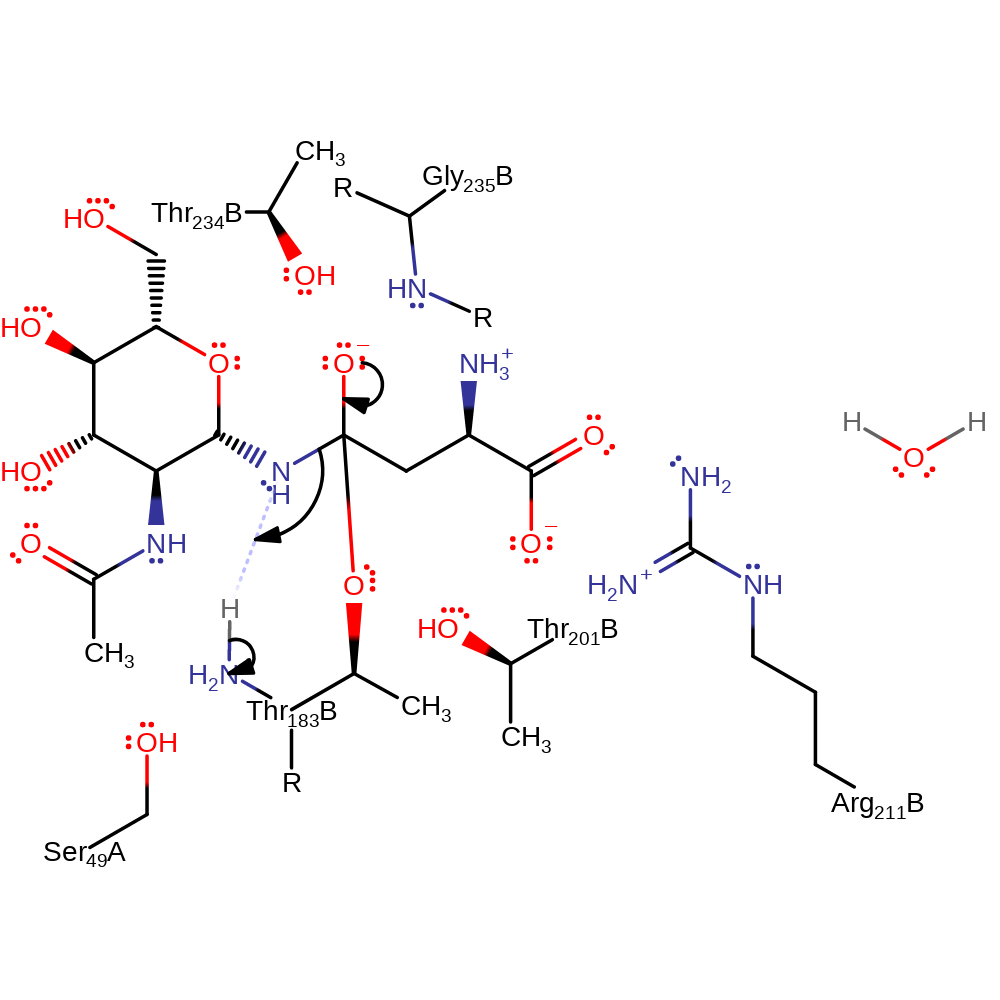
Step 2. The oxyanion initiates an elimination that cleaves the C-N bond, the sugar product deprotonates the N-terminus of Thr206
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly235(53)B (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Arg211(29)B | hydrogen bond acceptor, electrostatic stabiliser |
| Thr183(1)B | covalently attached, hydrogen bond acceptor |
| Ser49A | hydrogen bond acceptor, activator |
| Thr201(19)B | hydrogen bond donor |
| Thr234(52)B | electrostatic stabiliser |
| Thr183(1)B (N-term) | proton donor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, enzyme-substrate complex cleavage, intermediate collapse, intermediate formation, overall product formed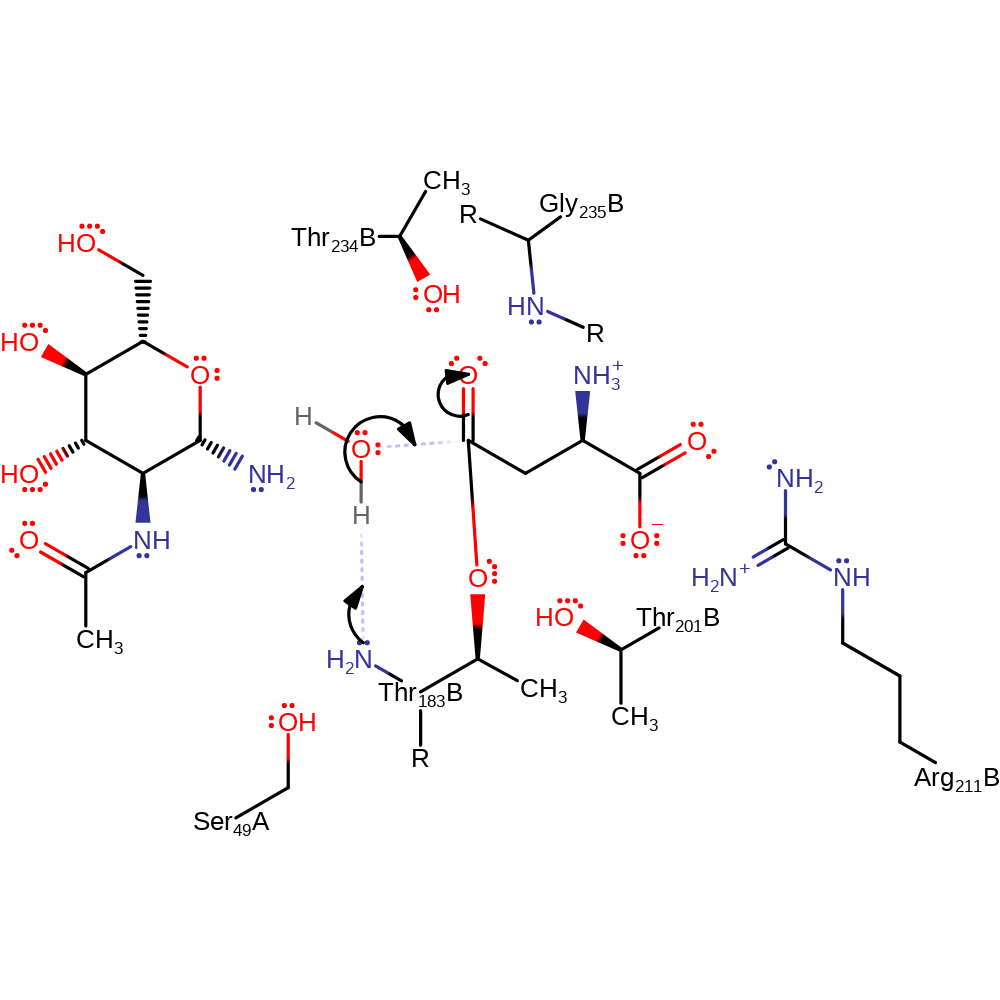
Step 3. The N-terminus of Thr206 deprotonates water, which attacks the carbonyl carbon of the covalently bound aspartate intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly235(53)B (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Arg211(29)B | hydrogen bond acceptor, electrostatic stabiliser |
| Thr183(1)B | covalently attached, hydrogen bond donor |
| Ser49A | hydrogen bond acceptor, activator |
| Thr201(19)B | hydrogen bond donor, activator |
| Thr234(52)B | electrostatic stabiliser |
| Thr183(1)B (N-term) | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation, intermediate formation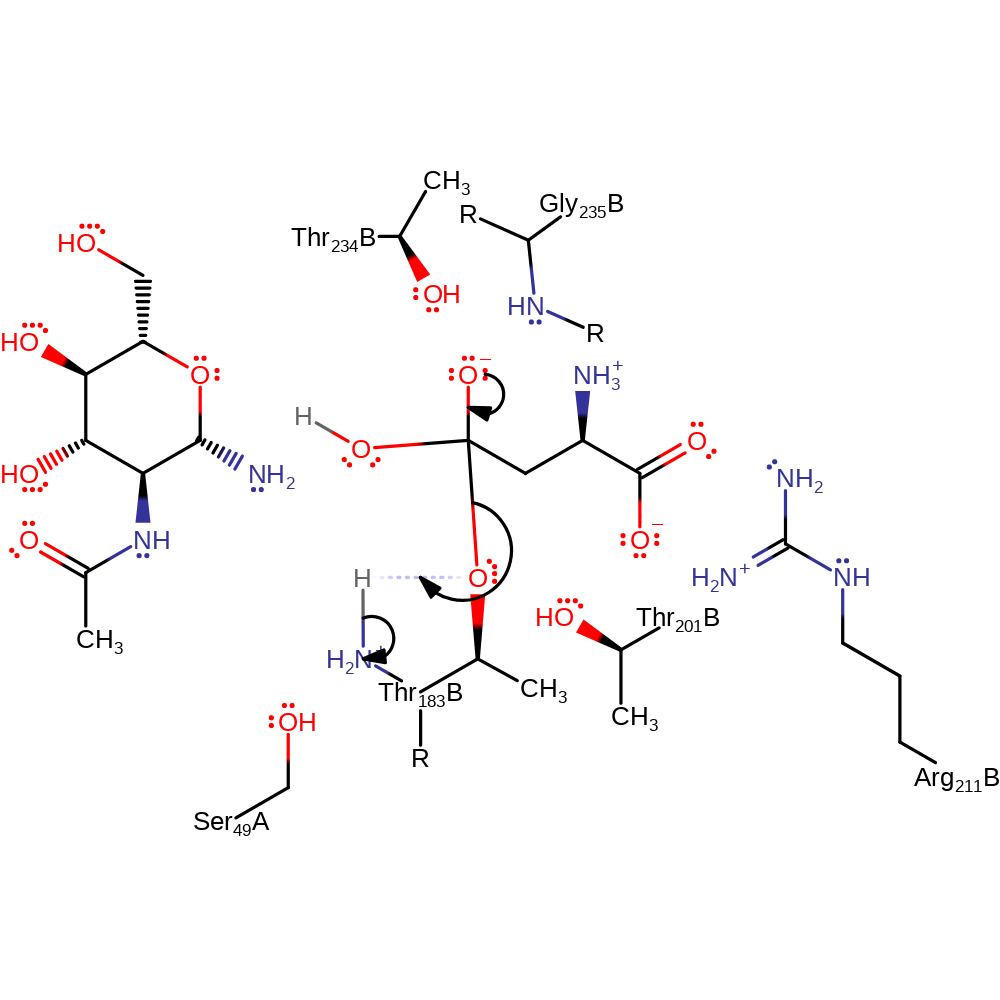
Step 4. The oxyanion initiates an elimination that cleaves the bond to the enzyme, the alcoholate of Thr206 deprotonates the N-terminus of Thr206
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly235(53)B (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Arg211(29)B | hydrogen bond acceptor, electrostatic stabiliser |
| Thr183(1)B | covalently attached, hydrogen bond acceptor |
| Ser49A | hydrogen bond acceptor, activator |
| Thr201(19)B | hydrogen bond donor |
| Thr234(52)B | electrostatic stabiliser |
| Thr183(1)B (N-term) | proton donor |
| Thr183(1)B | nucleofuge, proton acceptor |




 Download:
Download: