Oxalate decarboxylase
Oxalate decarboxylase (OxdC), isolated from Bacillus subtilus, catalyses the conversion of oxalate into formate and carbon dioxide. OxdC is part of the bicupin (contain two copies of the cupin domain) subset of the cupin superfamily. The enzymes requires Mn(II) and dioxygen as cofactors. The enzyme contains two manganese binding sites but it is currently thought that only the N-terminal site I possesses catalytic activity and that the C-terminal site II is structural. However, the matter is the subject of ongoing debate. Further, this enzyme contains an unusual cofactor in the form of molecular oxygen, which is thought to act as an activator to the Mn(II) centre by increasing its redox potential.
Reference Protein and Structure
- Sequence
-
O34714
 (4.1.1.2)
(4.1.1.2)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Bacillus subtilis subsp. subtilis str. 168 (Bacteria)

- PDB
-
1uw8
- CRYSTAL STRUCTURE OF OXALATE DECARBOXYLASE
(2.0 Å)



- Catalytic CATH Domains
-
2.60.120.10
 (see all for 1uw8)
(see all for 1uw8)
- Cofactors
- Manganese(2+) (1), Dioxygen (1) Metal MACiE
Enzyme Mechanism
Introduction
Mono-protonated oxalate binds to the Mn(II) ion in site I. Dioxygen coordinates to the metal to form a Mn(III)-superoxo species. The carboxylic acid of oxalate is deprotonated by Glu162 with concomitant electron transfer to Mn(III) to form the Mn(II)-bound oxalate radical anion. This intermediate decarboxylates to form the Mn(II)-bound formyl carbon-centred radical intermediate. This intermediate is then protonated by Glu162 with concomitant electron transfer from Mn(II) to form Mn(III)-bound formate. Formate and dioxygen dissociate to leave the enzyme in the resting state.
Catalytic Residues Roles
| UniProt | PDB* (1uw8) | ||
| Arg92 | Arg92A | Arg92 polarises the Mn-bound carbonyl group of formate, which increases the rate of decarboxylation. It might also stabilise a negative charge supported by this oxygen during the reaction. | attractive charge-charge interaction, hydrogen bond donor, electrostatic stabiliser |
| His95, His97, Glu101, His140 | His95A, His97A, Glu101A, His140A | Forms part of the catalytic manganese binding site. | metal ligand |
| Glu162 | Glu162A | Glu162 removes a proton from the carboxylic acid of Mn(III)-bound oxalate leading to the formation of the Mn(II)-bound oxalate radical anion intermediate. After decarboxylation it donates the proton to the Mn(II)-bound formyl radical to produce Mn(III)-bound formate. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, electrostatic stabiliser |
Chemical Components
coordination, intermediate formation, coordination to a metal ion, overall reactant used, colligation, radical termination, cofactor used, proton transfer, redox reaction, radical formation, bond polarisation, unimolecular elimination by the conjugate base, radical propagation, decarboxylation, overall product formed, electron transfer, homolysis, heterolysis, native state of cofactor regenerated, decoordination from a metal ion, intermediate terminated, inferred reaction stepReferences
- Svedruzić D et al. (2007), Arch Biochem Biophys, 464, 36-47. Investigating the roles of putative active site residues in the oxalate decarboxylase from Bacillus subtilis. DOI:10.1016/j.abb.2007.03.016. PMID:17459326.
- Campomanes P et al. (2014), J Am Chem Soc, 136, 2313-2323. Assigning the EPR fine structure parameters of the Mn(II) centers in Bacillus subtilis oxalate decarboxylase by site-directed mutagenesis and DFT/MM calculations. DOI:10.1021/ja408138f. PMID:24444454.
- Saylor BT et al. (2012), Biochemistry, 51, 2911-2920. A structural element that facilitates proton-coupled electron transfer in oxalate decarboxylase. DOI:10.1021/bi300001q. PMID:22404040.
- Angerhofer A et al. (2007), J Phys Chem B, 111, 5043-5046. Multifrequency EPR Studies on the Mn(II) Centers of Oxalate Decarboxylase. DOI:10.1021/jp0715326. PMID:17444678.
- Just VJ et al. (2007), Biochem J, 407, 397-406. The identity of the active site of oxalate decarboxylase and the importance of the stability of active-site lid conformations1. DOI:10.1042/bj20070708. PMID:17680775.
- Muthusamy M et al. (2006), Biochemistry, 45, 10667-10673. Real-Time Monitoring of the Oxalate Decarboxylase Reaction and Probing Hydron Exchange in the Product, Formate, Using Fourier Transform Infrared Spectroscopy†. DOI:10.1021/bi060460q. PMID:16939218.
- Just VJ et al. (2004), J Biol Chem, 279, 19867-19874. A Closed Conformation ofBacillus subtilisOxalate Decarboxylase OxdC Provides Evidence for the True Identity of the Active Site. DOI:10.1074/jbc.m313820200. PMID:14871895.
- Whittaker MM et al. (2002), J Biol Inorg Chem, 7, 136-145. Characterization of recombinant barley oxalate oxidase expressed by Pichia pastoris. DOI:10.1007/s007750100281. PMID:11862550.
- Anand R et al. (2002), Biochemistry, 41, 7659-7669. Structure of Oxalate Decarboxylase fromBacillus subtilisat 1.75 Å Resolution†,‡. DOI:10.1021/bi0200965.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu162A | hydrogen bond acceptor |
| Arg92A | hydrogen bond donor |
| His95A | metal ligand |
| His140A | metal ligand |
| Glu101A | metal ligand |
| His97A | metal ligand |
Chemical Components
coordination, intermediate formation, coordination to a metal ion, overall reactant usedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu162A | hydrogen bond acceptor, electrostatic stabiliser |
| Arg92A | hydrogen bond donor, electrostatic stabiliser |
| His95A | metal ligand |
| His140A | metal ligand |
| Glu101A | metal ligand |
| His97A | metal ligand |
Chemical Components
colligation, radical termination, cofactor used, intermediate formation
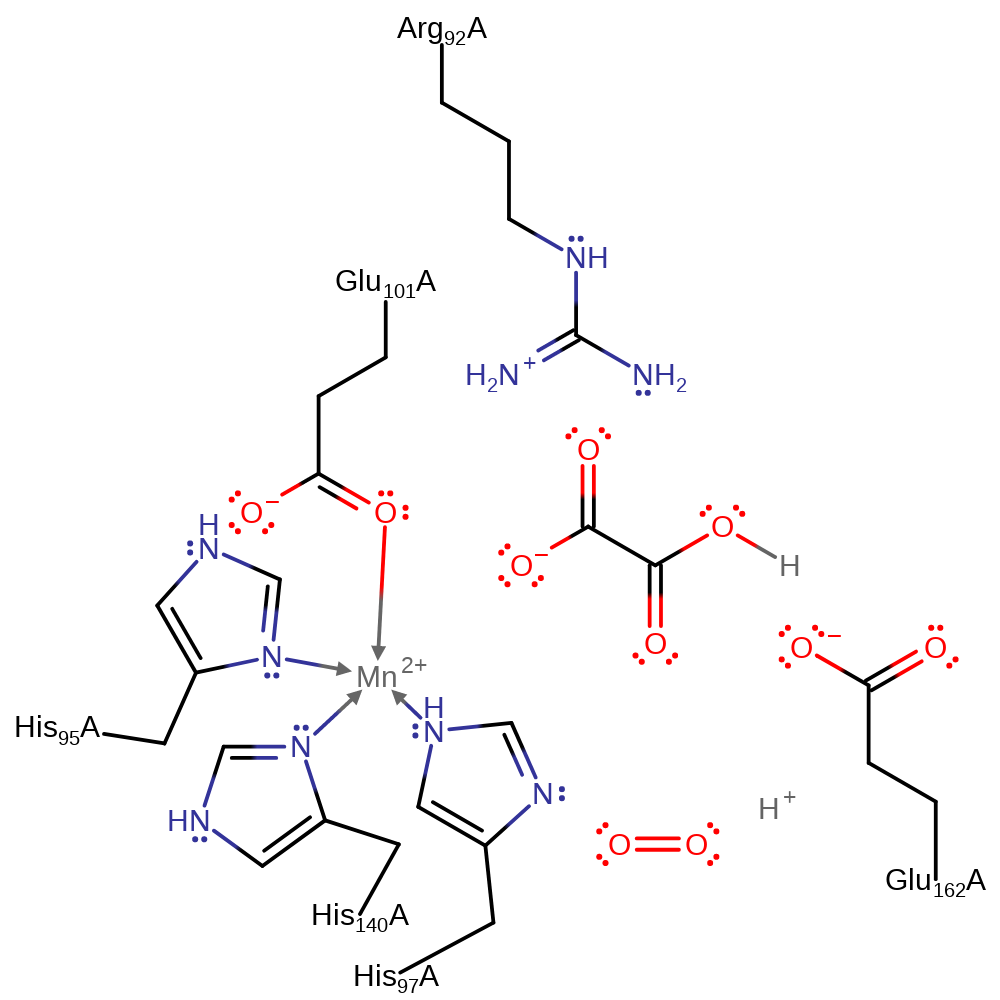
Step 3. Glu162 activates the Mn(III)-bound oxalate through deprotonation. This causes the oxalate bridging oxygen to transfer an electron to the Mn(III) centre. Deprotonation of oxalate is accompanied by electron transfer from the bridging oxygen to the manganese centre. It should be noted that the oxalate radical exists as a hybrid of two resonance forms: with the charges of the carbonyl separated and with the carbonyl as normal. The former is more favoured (70:30) and has been shown as such.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu162A | hydrogen bond acceptor |
| Arg92A | attractive charge-charge interaction, hydrogen bond donor, electrostatic stabiliser |
| His95A | metal ligand |
| His140A | metal ligand |
| Glu101A | metal ligand |
| His97A | metal ligand |
| Glu162A | proton acceptor |
Chemical Components
proton transfer, redox reaction, radical formation, bond polarisation, intermediate formationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu162A | hydrogen bond donor |
| Arg92A | electrostatic stabiliser, attractive charge-charge interaction, hydrogen bond donor |
| His95A | metal ligand |
| His140A | metal ligand |
| Glu101A | metal ligand |
| His97A | metal ligand |
Chemical Components
ingold: unimolecular elimination by the conjugate base, radical propagation, decarboxylation, intermediate formation, overall product formed
Step 5. The Mn(II) centre transfers an electron to the formyl intermediate carbon. The newly formed carbanion then deprotonates Glu162.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu162A | hydrogen bond donor |
| Arg92A | hydrogen bond donor, electrostatic stabiliser |
| His95A | metal ligand |
| His140A | metal ligand |
| Glu101A | metal ligand |
| His97A | metal ligand |
| Glu162A | proton donor |
Chemical Components
electron transfer, proton transfer, radical propagation, radical termination, intermediate formation
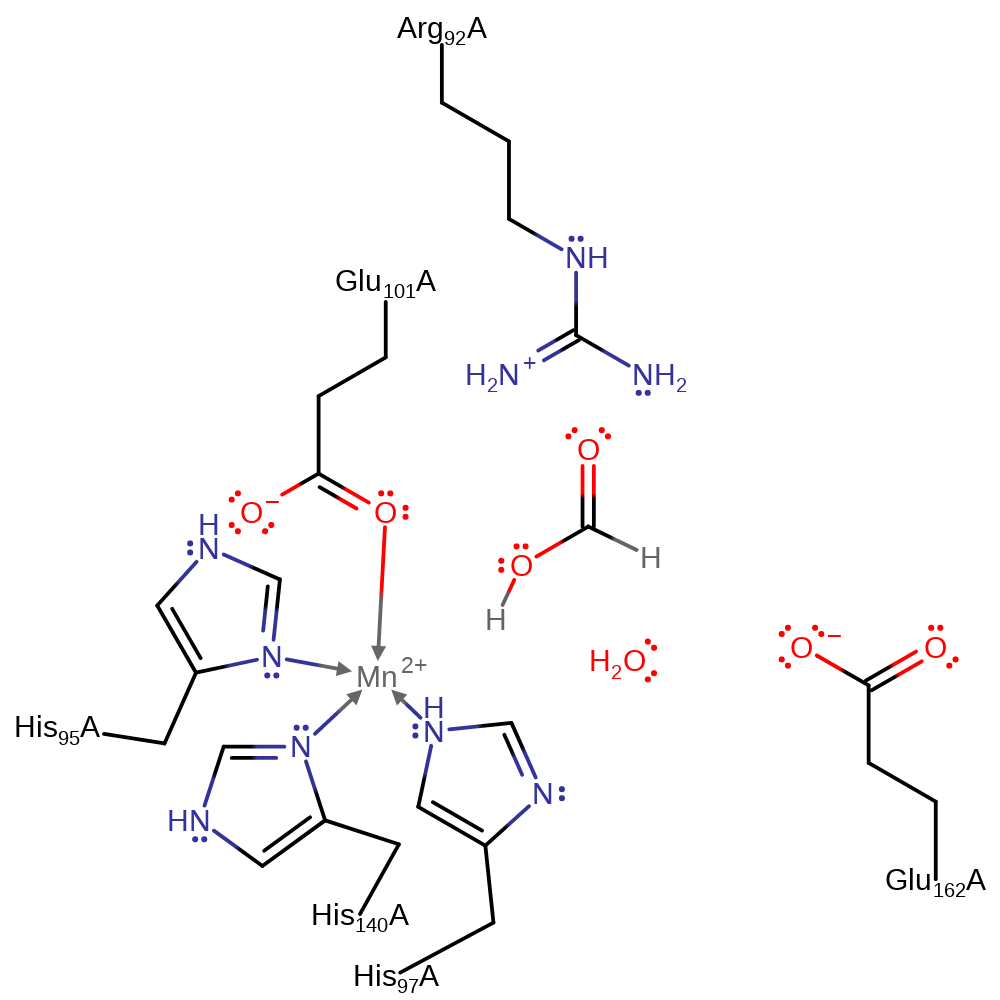
Step 6. Oxygen dissociates from the Mn(III) centre, regenerating the Mn(II) centre. It is unclear whether the oxygen cofactor or the formate product dissociates first. However, the dissociation of the oxygen cofactor results in the regeneration of the Mn(II) centre. For simplicity, both dissociation events are shown in a single step.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg92A | hydrogen bond donor |
| His95A | metal ligand |
| His140A | metal ligand |
| Glu101A | metal ligand |
| His97A | metal ligand |
Chemical Components
homolysis, heterolysis, radical formation, radical propagation, native state of cofactor regenerated, decoordination from a metal ion, intermediate terminated, overall product formedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His95A | metal ligand |
| His140A | metal ligand |
| Glu101A | metal ligand |
| His97A | metal ligand |







 Download:
Download: