Proteasome endopeptidase complex
The 26S proteasome is the central enzyme of non-lysosomal protein degradation. It is involved in the removal of misfolded or incorrectly assembled proteins, and also in the degradation of short lived regulatory proteins including transcription factors and the cyclins of cell-cycle control. The catalytic core of the complex is formed by the 20S proteasome, which has the form of a barrel-shaped particle composed of four stacked seven-membered rings. In yeast and higher eukaryotes, the rings are made up of 14 different but related subunits, the overall complex containing two subunits of each type. These can be classified into two families, alpha-type and beta-type. The beta type subunits form the inner two rings of the complex and at least three of them (beta-1, beta-2 and beta-5) are catalytically active. The cleavage specificities of these sites are determined largely by their S1 pockets and the three major specificities of the proteasome - peptidylglutamil-hydrolysing, trypsin-like, and chymotrypsin-like - have been assigned respectively to beta-1, beta-2 and beta-5.
The proteasome from archaea such as Thermoplasma acidophilum is simpler, consisting of 14 copies each of only two different subunits, alpha and beta. The archaeal enzyme has only a single, chymotrypsin-like activity although it has been shown to hydrolyse almost any peptide bond in denatured substrates.
In all cases the beta subunits are synthesised as inactive precursors which undergo autocatalytic cleavage to expose the catalytically active N-terminal residue.
Reference Protein and Structure
- Sequence
-
P25043
 (3.4.25.1)
(3.4.25.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Saccharomyces cerevisiae S288c (Baker's yeast)

- PDB
-
1ryp
- CRYSTAL STRUCTURE OF THE 20S PROTEASOME FROM YEAST AT 2.4 ANGSTROMS RESOLUTION
(1.9 Å)



- Catalytic CATH Domains
-
3.60.20.10
 (see all for 1ryp)
(see all for 1ryp)
Enzyme Reaction (EC:3.4.25.1)
Enzyme Mechanism
Introduction
The proteasome is an N-terminal nucleophilic (Ntn) hydrolase. The hydroxyl group of the N-terminal threonine residue is deprotonated by the terminal -NH2 group as it attacks the substrate carbonyl to form a tetrahedral intermediate. This intermediate is stabilised by an oxyanion hole formed from the backbone NH group of Gly 47. Collapse of the tetrahedral intermediate with protonation of the departing amine group by the terminal -NH3+ group generates an acyl enzyme intermediate; this is then attacked by a water molecule that is deprotonated by the terminal -NH2 group of Thr 1 acting again as a general base. The nearby Lys 33 (in its protonated form) functions to interact with the O-gamma of Thr 1 and provide a positive charge to promote deprotonation of this group in the first step of the reaction. The autocatalytic processing reaction also involves nucleophilic attack by the threonine OH, although there is not yet an amine base present to remove the proton. The oxyanion hole for the autocatalytic reaction is provided by Ser 129.
Catalytic Residues Roles
| UniProt | PDB* (1ryp) | ||
| Gly76 (main-N), Arg48 (main-C) | Gly47I (main-N), Arg19I (main-C) | Forms oxyanion hole in substrate cleavage reactions; this stabilises the tetrahedral intermediate and the transition state leading to it. | electrostatic stabiliser |
| Thr30 (N-term) | Thr1I (N-term) | N terminus functions to remove the proton from the side chain oxygen and then to protonate the amine leaving group from the tetrahedral intermediate. Later it deprotonates a water molecule which attacks the acyl enzyme intermediate | proton acceptor, proton donor |
| Lys62 | Lys33I | The positive charge on Lys33 may shift the intrinsic pKa of the catalytic water molecule and of the Thr1 amino and hydroxyl groups, enhancing their nucleophilicity | activator, electrostatic stabiliser |
| Thr30 | Thr1I | Side chain oxygen acts as a nucleophile to attack the peptide carbonyl and form an acyl-enzyme intermediate. | covalently attached, hydrogen bond acceptor, hydrogen bond donor, nucleofuge, proton acceptor, nucleophile, proton donor |
| Asp46 | Asp17I | Asp17 orients Lys33 in analogy to the role of Asp in chymotrypsin [PMID:7725097] | activator, steric locator |
| Ser158, Asp195, Ser198 | Ser129I, Asp166I, Ser169I | Forms oxyanion hole (using backbone NH and side chain OH) in the autocatalytic cleavage reaction. Both these residues are close to Thr1 N, and both hydroxyl groups are hydrogen-bonded to Asp166. Together these residues help orient the N-terminus group to enable it to act as a general acid/base. | activator, electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation, intermediate formation, unimolecular elimination by the conjugate base, enzyme-substrate complex cleavage, intermediate collapse, overall product formed, intermediate terminated, native state of enzyme regeneratedReferences
- Löwe J et al. (1995), Science, 268, 533-539. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. DOI:10.1126/science.7725097. PMID:7725097.
- Wei D et al. (2013), J Phys Chem B, 117, 13418-13434. Fundamental reaction pathway for peptide metabolism by proteasome: insights from first-principles quantum mechanical/molecular mechanical free energy calculations. DOI:10.1021/jp405337v. PMID:24111489.
- Groll M et al. (2005), Chembiochem, 6, 222-256. Molecular Machines for Protein Degradation. DOI:10.1002/cbic.200400313. PMID:15678420.
- Groll M et al. (2003), J Mol Biol, 327, 75-83. Investigations on the Maturation and Regulation of Archaebacterial Proteasomes. DOI:10.1016/s0022-2836(03)00080-9. PMID:12614609.
- Oinonen C et al. (2000), Protein Sci, 9, 2329-2337. Structural comparison of Ntn-hydrolases. DOI:10.1110/ps.9.12.2329. PMID:11206054.
- Groll M et al. (1999), Proc Natl Acad Sci U S A, 96, 10976-10983. The catalytic sites of 20S proteasomes and their role in subunit maturation: A mutational and crystallographic study. DOI:10.1073/pnas.96.20.10976. PMID:10500111.
- Ditzel L et al. (1998), J Mol Biol, 279, 1187-1191. Conformational constraints for protein self-cleavage in the proteasome. DOI:10.1006/jmbi.1998.1818. PMID:9642094.
- Groll M et al. (1997), Nature, 386, 463-471. Structure of 20S proteasome from yeast at 2.4Å resolution. DOI:10.1038/386463a0. PMID:9087403.
- Seemuller E et al. (1996), Nature, 382, 468-470. Autocatalytic processing of the 20S proteasome. DOI:10.1038/382468a0. PMID:8684489.
- Seemüller E et al. (1995), Science, 268, 579-582. Proteasome from Thermoplasma acidophilum: a threonine protease. DOI:10.1126/science.7725107. PMID:7725107.
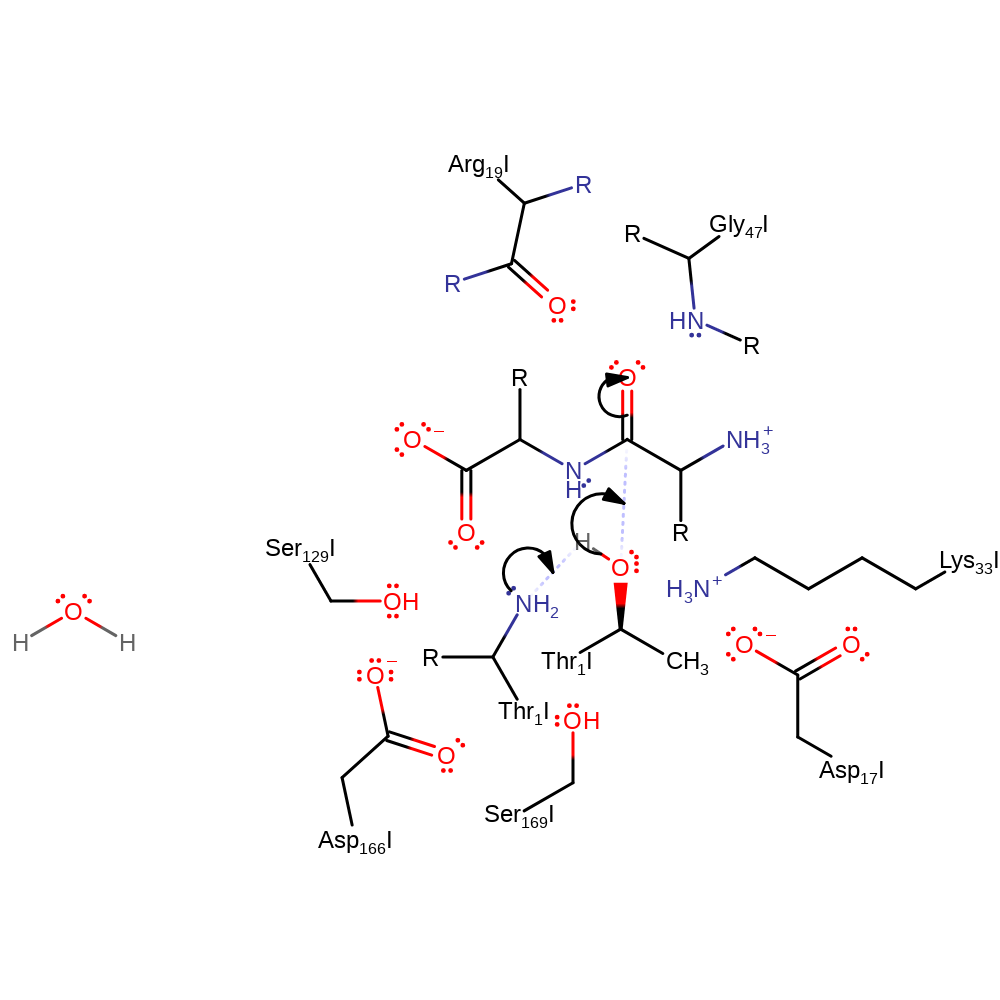
Step 1. The N-terminus of Thr1 deprotonates the alcohol group of Thr1, which attacks the peptide bond in a nucleophilic addition.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr1I | hydrogen bond donor, hydrogen bond acceptor |
| Arg19I (main-C) | activator |
| Lys33I | activator |
| Ser129I | activator |
| Asp166I | activator |
| Ser169I | activator |
| Gly47I (main-N) | electrostatic stabiliser |
| Ser129I | electrostatic stabiliser |
| Asp17I | steric locator |
| Asp166I | steric locator |
| Thr1I (N-term) | proton acceptor |
| Thr1I | nucleophile, proton donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation, intermediate formation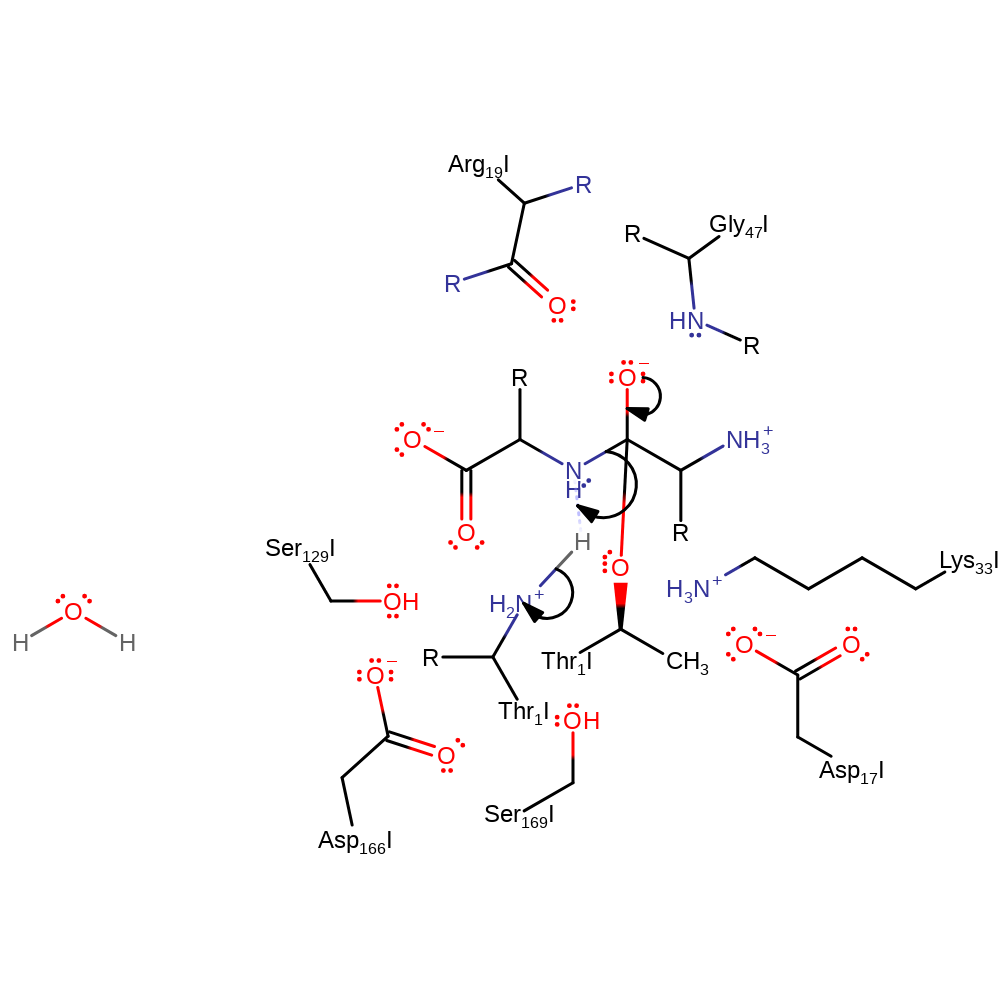
Step 2. The oxyanion initiates an elimination that cleaves the peptide bond, the N-terminal product deprotonates the N-terminus of Thr1
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr1I | covalently attached |
| Gly47I (main-N) | electrostatic stabiliser |
| Ser129I | electrostatic stabiliser |
| Ser169I | electrostatic stabiliser |
| Asp166I | steric locator, activator |
| Asp17I | steric locator |
| Thr1I (N-term) | proton donor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, enzyme-substrate complex cleavage, intermediate collapse, intermediate formation, overall product formed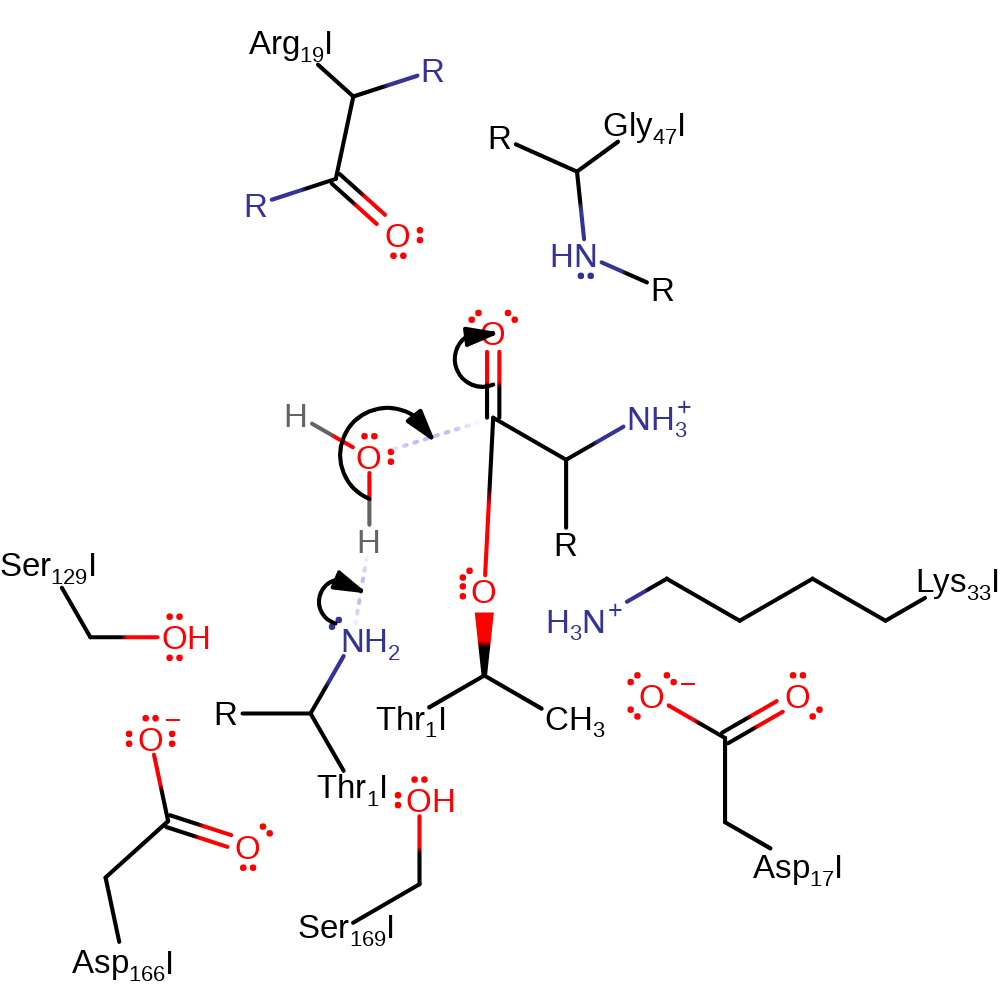
Step 3. The N-terminus of Thr1 deprotonates water, which attacks the carbonyl carbon of the covalently bound C-terminal intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr1I | covalently attached |
| Gly47I (main-N) | electrostatic stabiliser |
| Ser129I | electrostatic stabiliser |
| Ser169I | electrostatic stabiliser |
| Arg19I (main-C) | activator |
| Lys33I | activator |
| Ser129I | activator |
| Ser169I | activator |
| Asp17I | steric locator |
| Asp166I | steric locator |
| Asp17I | activator |
| Asp166I | activator |
| Thr1I (N-term) | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation, intermediate formation
Step 4. The oxyanion initiates an elimination that cleaves the peptide bond, the alcoholate of Thr1 deprotonates the N-terminus of Thr1
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr1I | hydrogen bond acceptor |
| Asp17I | steric locator |
| Asp166I | steric locator, activator |
| Lys33I | electrostatic stabiliser |
| Gly47I (main-N) | electrostatic stabiliser |
| Ser129I | electrostatic stabiliser |
| Ser169I | electrostatic stabiliser |
| Thr1I (N-term) | proton donor |
| Thr1I | proton acceptor, nucleofuge |



 Download:
Download: