Ribonuclease HI
Members of the RNase H family hydrolyze the P-O3' bond of the RNA strand of an RNA-DNA hybrid duplex in the presence of divalent cations such as Mg(II) and Mn(II). RNase HI is found in Escherichia coli and is structurally homologous to the RNase H domain of HIV-1 reverse transcriptase, a target for anti-HIV therapy.
Two alternative mechanisms have been proposed for this enzyme. One is a two-metal-ion mechanism where one of the two metal ions activates the attacking hydroxide ion and the other is a general acid-base mechanism where an amino acid residue fulfils this role. Computational studies seem to favour the two metal ion proposal, however NMR and kinetic studies suggest only one metal binds to this protein and that the protein is inactivated by subsequent metal binding events.
Reference Protein and Structure
- Sequence
-
P0A7Y4
 (3.1.26.4)
(3.1.26.4)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1rdd
- CRYSTAL STRUCTURE OF ESCHERICHIA COLI RNASE HI IN COMPLEX WITH MG2+ AT 2.8 ANGSTROMS RESOLUTION: PROOF FOR A SINGLE MG2+ SITE
(2.8 Å)



- Catalytic CATH Domains
-
3.30.420.10
 (see all for 1rdd)
(see all for 1rdd)
- Cofactors
- Magnesium(2+) (1), Water (1) Metal MACiE
Enzyme Reaction (EC:3.1.26.4)
Enzyme Mechanism
Introduction
The basic mechanism is SN2-like inline displacement of the 3' end of the scissile phosphate group by a water nucleophile in a single step, passing through a pentavalent transition state. The water nucleophile is held and activated by His 124, which deprotonates the water molecule, enabling attack as a hydroxide ion. His 124 itself is made more basic by hydrogen bonding to the substrate phosphate group 3' to the scissile phosphodiester bond. The 3' hydroxyl leaving group is activated as a leaving group by protonation by a water molecule; the water molecule itself is acidified by the Mg(II) cation and the 2' hydroxyl of the substrate ribose 5' to the scissile phosphodiester bond. The residues Asp 134 and Glu 48 are important for orienting the water molecules.
Catalytic Residues Roles
| UniProt | PDB* (1rdd) | ||
| Glu48 | Glu48A | Glu48 anchors the water molecule that acts as a general acid and helps activate it. | increase basicity, increase acidity |
| Asp134 | Asp134A | Asp134 holds the nucleophilic water molecule in place and helps activate it. | increase acidity |
| His124 | His124A | Activates the water nucleophile by deprotonation. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Asp10, Gly11 (main-C), Asp70 | Asp10A, Gly11A (main-C), Asp70A | Forms part of the magnesium binding site. | metal ligand |
Chemical Components
proton transfer, bimolecular nucleophilic substitution, overall reactant used, overall product formed, hydrolysis, native state of enzyme regenerated, inferred reaction stepReferences
- Haruki M et al. (2000), Biochemistry, 39, 13939-13944. Catalysis by Escherichia coli ribonuclease HI is facilitated by a phosphate group of the substrate. DOI:10.1021/bi001469+. PMID:11076536.
- Haruki M et al. (1994), Eur J Biochem, 220, 623-631. Investigating the role of conserved residue Asp134 in Escherichia coli ribonuclease HI by site-directed random mutagenesis. DOI:10.1111/j.1432-1033.1994.tb18664.x. PMID:8125123.
- Katayanagi K et al. (1993), Proteins, 17, 337-346. Crystal structure ofEscherichia coli RNase HI in complex with Mg2+ at 2.8 Å resolution: Proof for a single Mg2+-binding site. DOI:10.1002/prot.340170402. PMID:8108376.
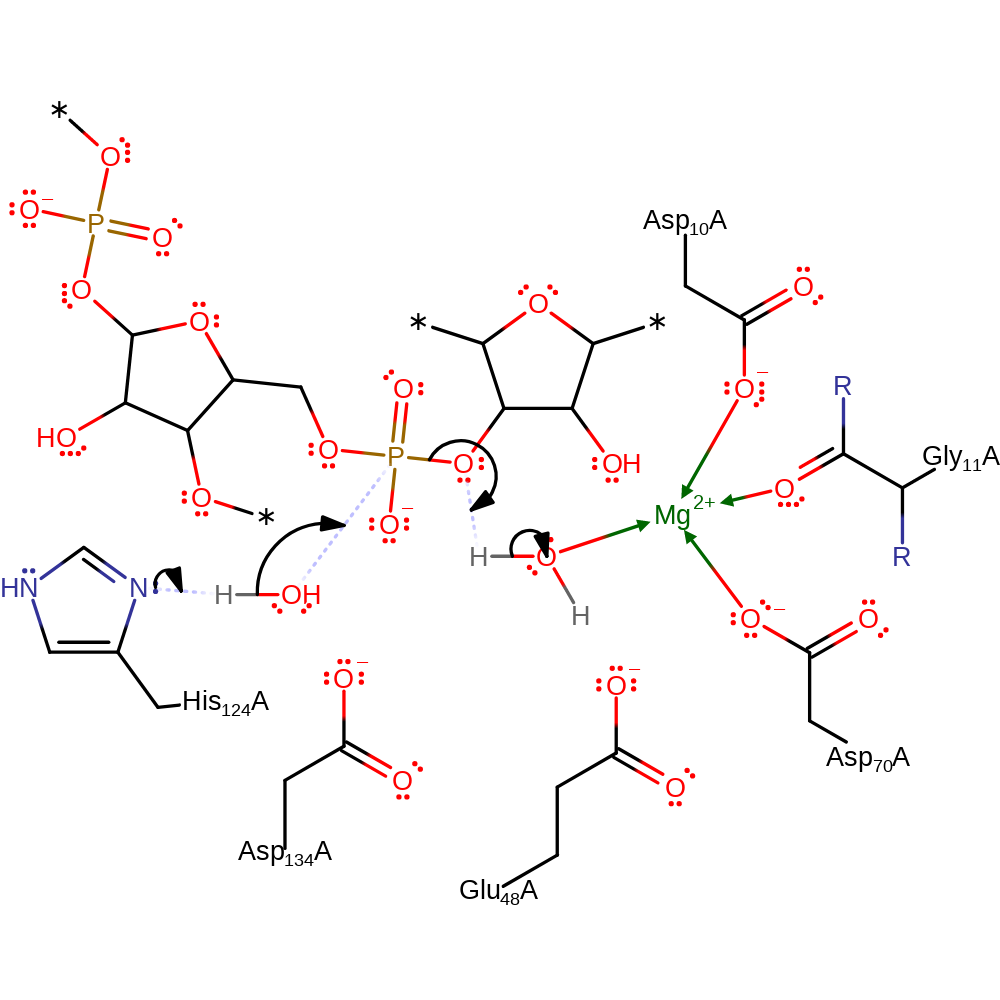
Step 1. His124 deprotonates water, which attacks the phosphate of RNA in a nucleophilic substitution that results in the cleavage of the phosphate bond, the 5' end of the RNA molecule reprotonates from magnesium activated water. Reaction proceeds through a pentavalent transition state. The pro-RP-oxygen of the phosphate group 3' to the scissile phosphodiester bond contributes to orient His124 to the best position for catalytic function through the formation of a hydrogen bond [PMID:11076536].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His124A | hydrogen bond donor, hydrogen bond acceptor |
| Asp10A | metal ligand |
| Gly11A (main-C) | metal ligand |
| Asp70A | metal ligand |
| Glu48A | increase acidity |
| Asp134A | increase acidity |
| His124A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, overall reactant used, overall product formed, hydrolysis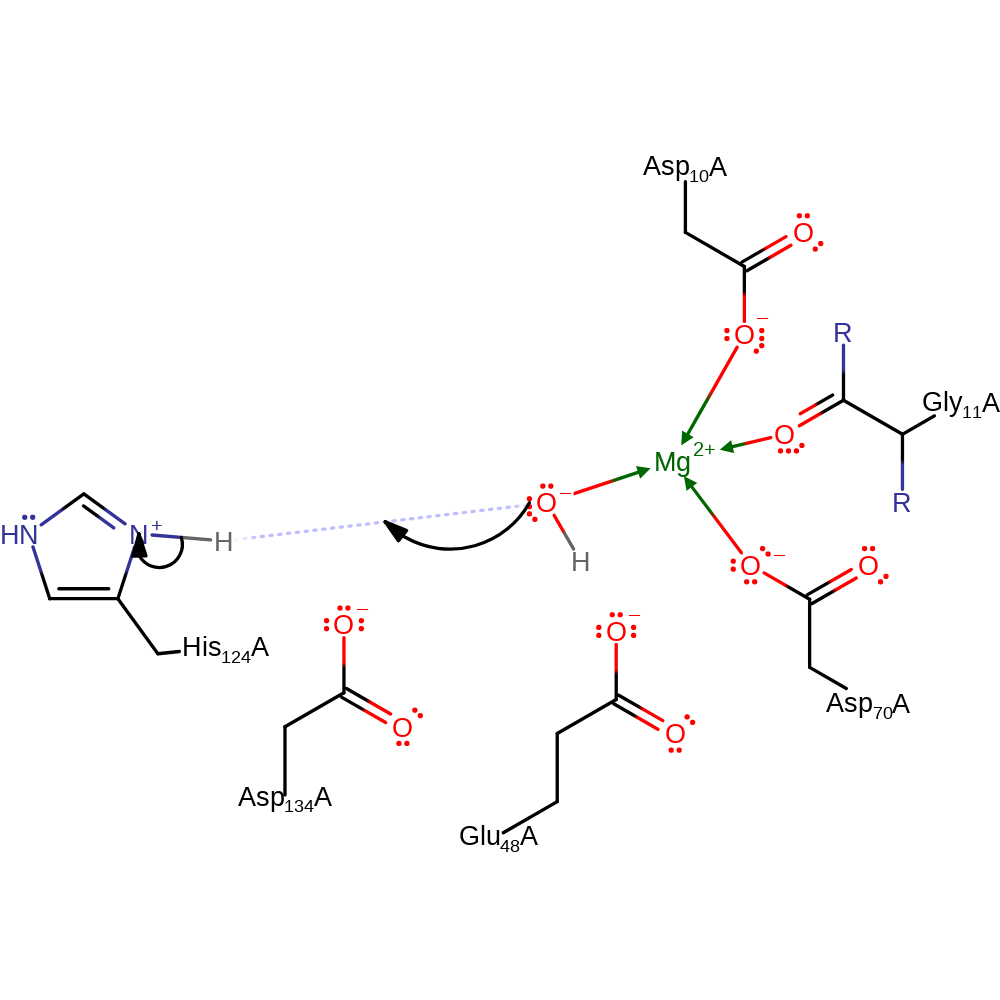
Step 2. Water deprotonates His124, regenerating the enzyme active site.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His124A | hydrogen bond donor |
| Asp10A | metal ligand |
| Gly11A (main-C) | metal ligand |
| Asp70A | metal ligand |
| Glu48A | increase basicity |
| His124A | proton donor |
Chemical Components
proton transfer, native state of enzyme regenerated, inferred reaction stepIntroduction
This proposal represents the two metal mechanism. Here the water molecules are activated by Mg(II). Computational studies propose that the reaction proceeds in two steps: (1) catalysed primarily by magnesium ion A and its ligands, a water molecule attacks the scissile phosphate. The transient phosphorane formed as a result of this nucleophilic attack decays by breaking the bond between the phosphate and the ribose oxygen. In the resulting intermediate, the dissociated but unprotonated leaving group forms an alkoxide coordinated to magnesium ion B. (2) The reaction is completed by protonation of the leaving group, with a neutral Asp132 as a likely proton donor.
Catalytic Residues Roles
| UniProt | PDB* (1rdd) | ||
| Asp10 | Asp10A | Acts as a bridging ligand between the two Mg(II) ions. | metal ligand |
| Asp134 | Asp134A | Forms part of the first magnesium binding site. Also thought to act as a general acid/base. It has been inferred that this residue starts in a negatively charged state and abstracts a proton from the nucleophilic water molecule, before donating the proton back to the final product. | metal ligand, proton acceptor, proton donor |
| Glu48, His124 | Glu48A, His124A | It is unclear what the function of these residues is in this proposal. | unknown |
| Gly11 (main-C), Asp70 | Gly11A (main-C), Asp70A | Forms part of the second magnesium binding site. | metal ligand |
Chemical Components
proton transfer, inferred reaction step, overall reactant used, overall product formed, hydrolysis, bimolecular nucleophilic substitution, native state of enzyme regeneratedReferences
- Nowotny M et al. (2005), Cell, 121, 1005-1016. Crystal Structures of RNase H Bound to an RNA/DNA Hybrid: Substrate Specificity and Metal-Dependent Catalysis. DOI:10.1016/j.cell.2005.04.024. PMID:15989951.
- Rosta E et al. (2011), J Am Chem Soc, 133, 8934-8941. Catalytic mechanism of RNA backbone cleavage by ribonuclease H from quantum mechanics/molecular mechanics simulations. DOI:10.1021/ja200173a. PMID:21539371.
- De Vivo M et al. (2008), J Am Chem Soc, 130, 10955-10962. Phosphodiester cleavage in ribonuclease H occurs via an associative two-metal-aided catalytic mechanism. DOI:10.1021/ja8005786. PMID:18662000.

Step 1. Asp134 abstracts a proton from the metal bound nucleophilic water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp10A | metal ligand |
| Gly11A (main-C) | metal ligand |
| Asp70A | metal ligand |
| Asp134A | metal ligand |
| Glu48A | unknown |
| His124A | unknown |
| Asp134A | proton acceptor |
Chemical Components
proton transfer, inferred reaction step, overall reactant used
Step 2. The catalytic water is activated by its position in the Mg(II) coordination sphere. The hydroxide attacks the phosphate of RNA in a nucleophilic substitution that results in the cleavage of the phosphate bond, the 5' end of the RNA molecule becomes part of the second Mg(II) coordination sphere. The reaction likely proceeds through a pentavalent transition state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu48A | unknown |
| His124A | unknown |
| Asp10A | metal ligand |
| Gly11A (main-C) | metal ligand |
| Asp70A | metal ligand |
| Asp134A | metal ligand |
Chemical Components
overall reactant used, overall product formed, hydrolysis, ingold: bimolecular nucleophilic substitution
Step 3. The alkoxide group abstracts a proton from Asp134 to form the second product of the reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu48A | unknown |
| His124A | unknown |
| Asp10A | metal ligand |
| Gly11A (main-C) | metal ligand |
| Asp70A | metal ligand |
| Asp134A | metal ligand |
| Asp134A | proton donor |




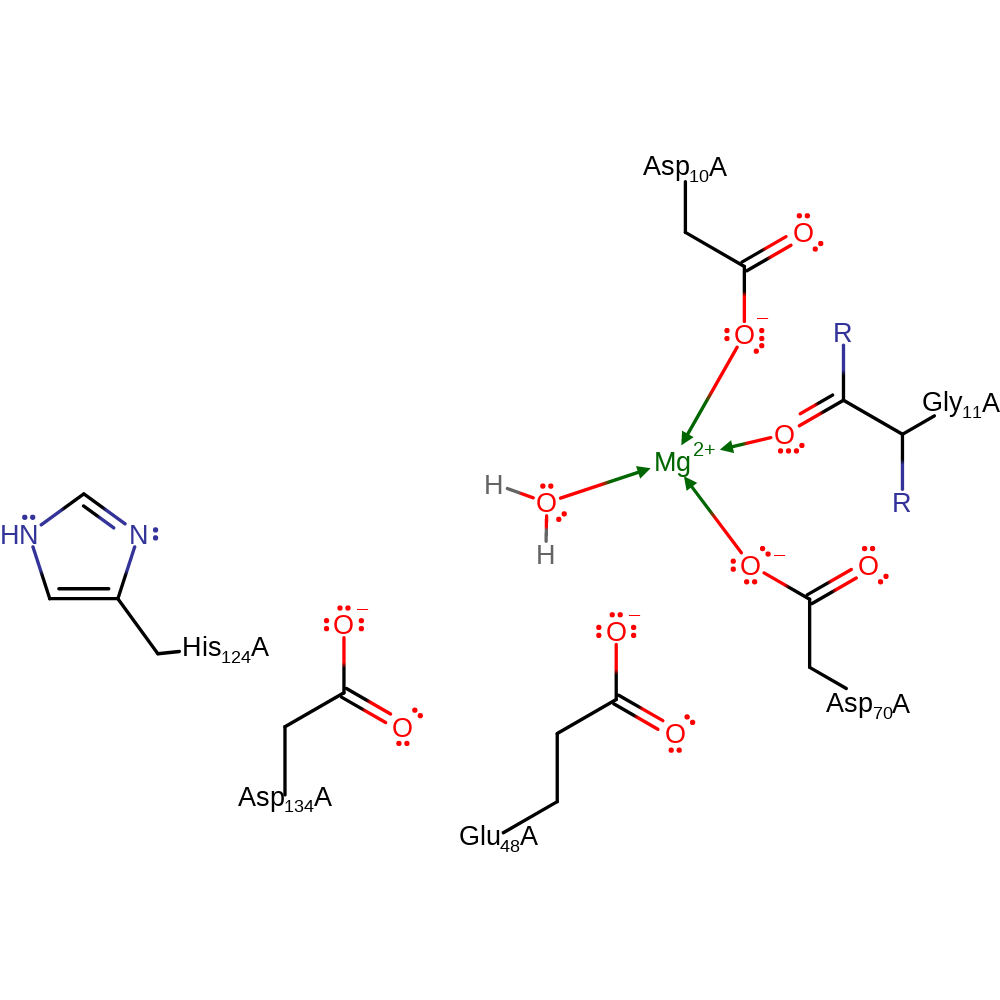 Download:
Download:  Download:
Download: