4-hydroxybenzoate 3-monooxygenase
4-hydroxybenzoate 3-monooxygenase is a flavoprotein that catalyses the incorporation of an atom of dioxygen into p-hydroxybenzoate (p-OHB) to form 3,4-dihydroxybenzoate (3,4DOHB). 3,4DOHB subsequently enters the beta-ketioadipate pathway of aromatic degradation, using molecular oxygen and NADPH.
The reaction occurs in two parts: reduction of the flavin adenine dinucleotide (FAD) in the enzyme by reduced nicotinamide adenine dinucleotide phosphate (NADPH) in response to binding p-hydroxybenzoate to the enzyme and oxidation of reduced FAD with oxygen to form a hydroperoxide, which then oxygenates p-hydroxybenzoate.
Reference Protein and Structure
- Sequence
-
P20586
 (1.14.13.2)
(1.14.13.2)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Pseudomonas aeruginosa PAO1 (Bacteria)

- PDB
-
1doc
- THE MOBIL FLAVIN OF 4-OH BENZOATE HYDROXYLASE: MOTION OF A PROSTHETIC GROUP REGULATES CATALYSIS
(2.0 Å)



- Catalytic CATH Domains
-
3.30.9.10
 3.50.50.60
3.50.50.60  (see all for 1doc)
(see all for 1doc)
- Cofactors
- Fadh2(2-) (1), Water (2)
Enzyme Reaction (EC:1.14.13.2)
Enzyme Mechanism
Introduction
The substrate is deprotonated via a proton relay chain from bulk solvent involving His72-water-water-Tyr385 and Tyr201. A conformational change allows NADP to bind, which initiates a reverse of the first proton transfer and a hydride transfer from NADP to the FAD cofactor. The FAD then undergoes a double bond rearrangement, resulting in the single electron transfer from FAD to dioxygen. The FAD and dioxygen radical species undergo a colligation reaction to form the FAD-peroxo adduct which then abstracts a proton from water. The 4-hydroxybenzoate substrate is deprotonated and then undergoes a double bond rearrangement that results in the ortho-position attacking the FAD-peroxo adduct in a nucleophilic substitution that cleaves the O-O bond. The FAD intermediate deprotonates the hydorxylated aromatic intermediate, cleaving the C-H bond and initiating a double bond rearrangement to yield 3,4-dihydroxybenzoic acid. This then takes a proton from water through the His72-water-water-Tyr385-Tyr201 proton transfer chain. Water deprotonates the 3,4-dihydroxybenzoic acid through the His72-water-water-Tyr385-Tyr201 proton transfer chain. Finally, the FAD-bound hydroxyl group initiates an intramolecular elimination of water, regenerating the FAD cofactor.
Catalytic Residues Roles
| UniProt | PDB* (1doc) | ||
| Tyr201, His72, Tyr385 | Tyr201A, His72A, Tyr385A | Form a hydrogen bonding network (with two water molecules) that links the active site with bulk solvent. This network forms a proton relay chain that functions throughout the reaction. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, proton relay |
| Pro293 (main-C), Lys297 | Pro293A (main-C), Lys297A | Act to bind and stabilise the reactive intermediates formed during the course of the later reaction steps. | hydrogen bond acceptor, electrostatic stabiliser |
Chemical Components
proton transfer, overall reactant used, intermediate formation, proton relay, aromatic unimolecular elimination by the conjugate base, aromatic bimolecular nucleophilic addition, hydride transfer, cofactor used, overall product formed, rate-determining step, electron transfer, radical formation, colligation, radical termination, bimolecular nucleophilic substitution, assisted keto-enol tautomerisation, intermediate terminated, aromatic intramolecular elimination, native state of cofactor regenerated, intermediate collapse, native state of enzyme regeneratedReferences
- Ortiz-Maldonado M et al. (2004), Biochemistry, 43, 15246-15257. Oxygen Reactions inp-Hydroxybenzoate Hydroxylase Utilize the H-Bond Network during Catalysis†. DOI:10.1021/bi048115t. PMID:15568817.
- Entsch B et al. (2005), Arch Biochem Biophys, 433, 297-311. Protein dynamics and electrostatics in the function of p-hydroxybenzoate hydroxylase. DOI:10.1016/j.abb.2004.09.029. PMID:15581585.
- Cole LJ et al. (2005), Biochemistry, 44, 8047-8058. Removal of a methyl group causes global changes in p-hydroxybenzoate hydroxylase. DOI:10.1021/bi050108x. PMID:15924424.
- Ortiz-Maldonado M et al. (2001), Biochemistry, 40, 8705-8716. Synergistic Interactions of Multiple Mutations on Catalysis during the Hydroxylation Reaction ofp-Hydroxybenzoate Hydroxylase: Studies of the Lys297Met, Asn300Asp, and Tyr385Phe Mutants Reconstituted with 8-Cl-Flavin†. DOI:10.1021/bi010892v. PMID:11467930.
- Eppink MH et al. (1999), FEBS Lett, 443, 251-255. Phe161 and Arg166 variants of p-hydroxybenzoate hydroxylase. Implications for NADPH recognition and structural stability. PMID:10025942.
- Ortiz-Maldonado M et al. (1999), Biochemistry, 38, 16636-16647. Structure-function correlations of the reaction of reduced nicotinamide analogues with p-hydroxybenzoate hydroxylase substituted with a series of 8-substituted flavins. PMID:10600126.
- Gatti DL et al. (1996), Biochemistry, 35, 567-578. pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis. DOI:10.1021/bi951344i. PMID:8555229.
- Lah MS et al. (1994), Biochemistry, 33, 1555-1564. Crystal structures of mutant Pseudomonas aeruginosa p-hydroxybenzoate hydroxylases: the Tyr201Phe, Tyr385Phe, and Asn300Asp variants. PMID:8312276.
- Entsch B et al. (1991), J Biol Chem, 266, 17341-17349. Catalytic function of tyrosine residues in para-hydroxybenzoate hydroxylase as determined by the study of site-directed mutants. PMID:1910043.
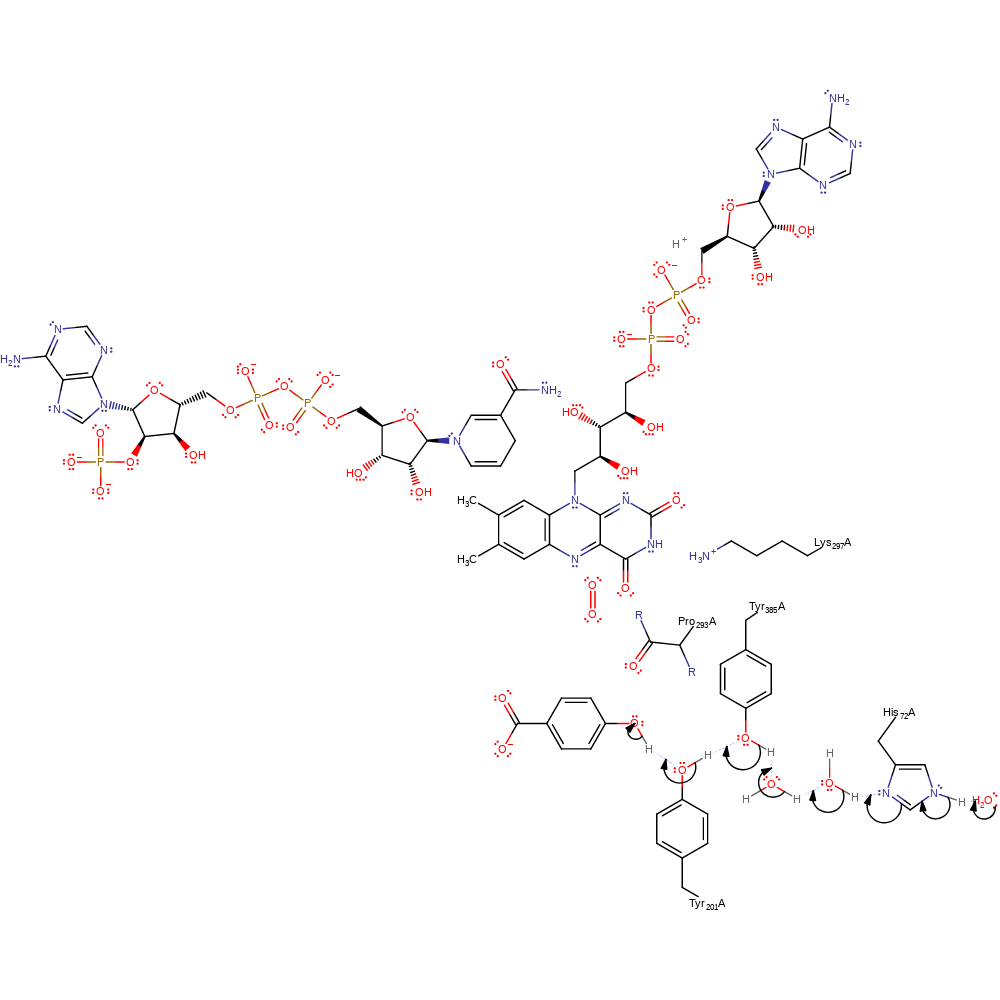
Step 1. In a proton transfer chain involving His72-water-water-Tyr385-Tyr201 water deprotonates the 4-hydroxybenzoate substrate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His72A | hydrogen bond acceptor, hydrogen bond donor, proton relay |
| Tyr201A | hydrogen bond donor, hydrogen bond acceptor, proton relay |
| Tyr385A | hydrogen bond donor, hydrogen bond acceptor, proton relay |
| Tyr201A | proton donor |
| His72A | proton donor, proton acceptor |
| Tyr201A | proton acceptor |
| Tyr385A | proton acceptor, proton donor |
Chemical Components
proton transfer, overall reactant used, intermediate formation, proton relay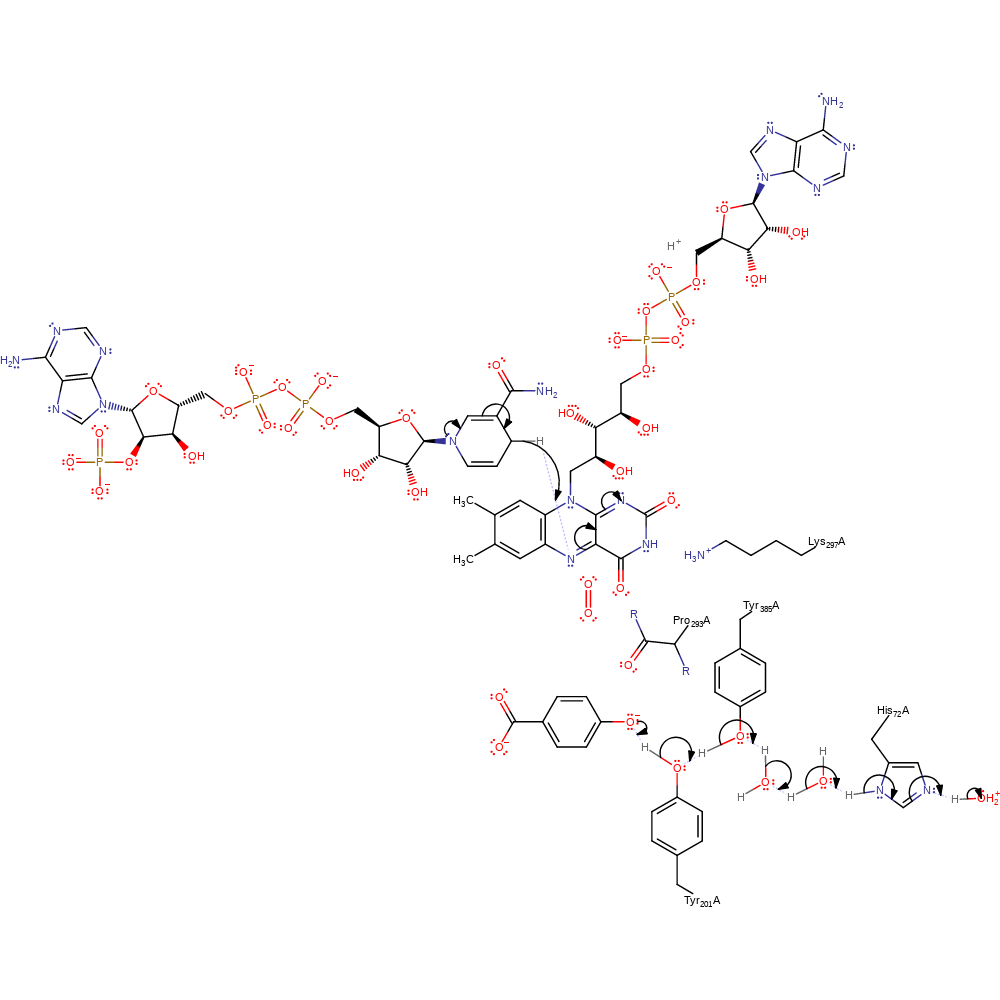
Step 2. A conformational change allows NADP to bind, which initiates a reverse of the first proton transfer and a hydride transfer from NADP to the FAD cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His72A | hydrogen bond acceptor, hydrogen bond donor, proton relay |
| Tyr201A | hydrogen bond donor, hydrogen bond acceptor, proton relay |
| Tyr385A | hydrogen bond donor, hydrogen bond acceptor, proton relay |
| Tyr385A | proton acceptor |
| His72A | proton donor |
| Tyr201A | proton donor |
| His72A | proton acceptor |
| Tyr385A | proton donor |
| Tyr201A | proton acceptor |
Chemical Components
ingold: aromatic unimolecular elimination by the conjugate base, ingold: aromatic bimolecular nucleophilic addition, hydride transfer, proton transfer, overall reactant used, cofactor used, intermediate formation, overall product formed, proton relay, rate-determining step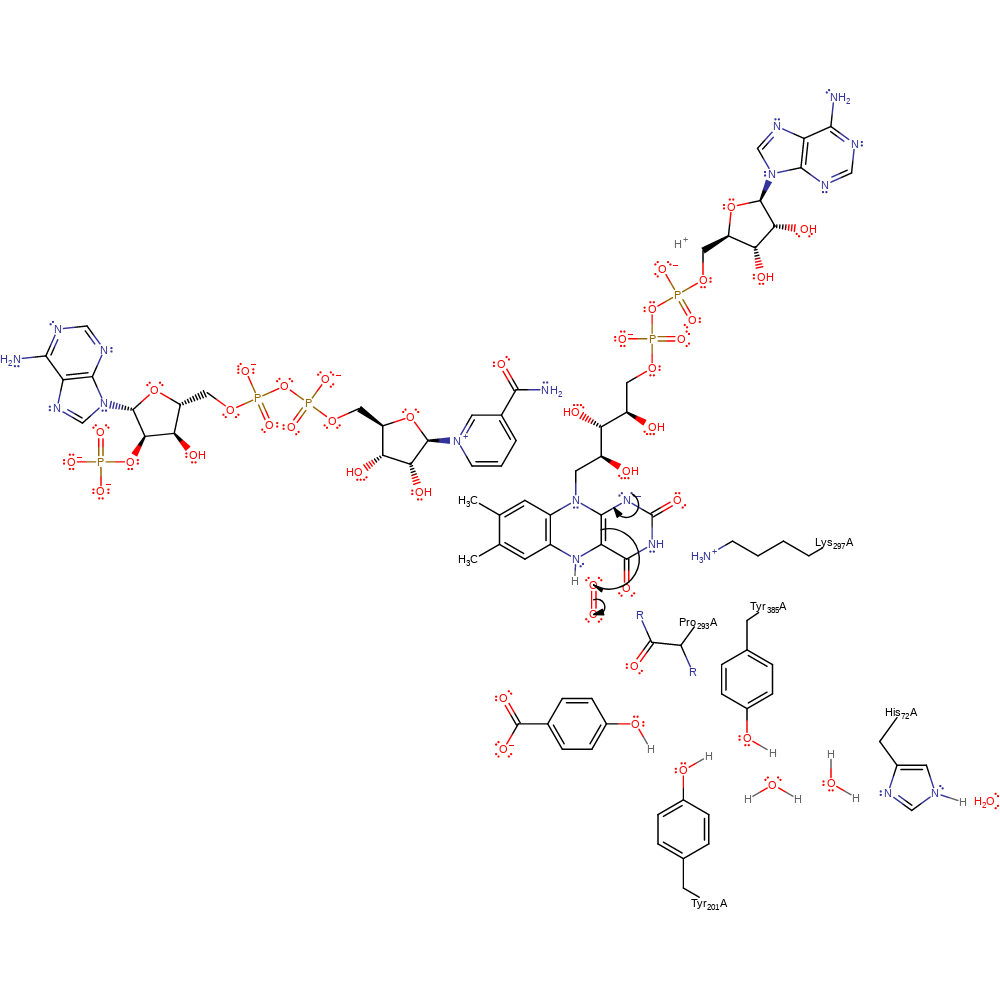
Step 3. FAD undergoes a double bond rearrangement that results in the single electron transfer from FAD to dioxygen.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His72A | hydrogen bond acceptor, hydrogen bond donor |
| Tyr201A | hydrogen bond acceptor, hydrogen bond donor |
| Tyr385A | hydrogen bond acceptor, hydrogen bond donor |
Chemical Components
electron transfer, radical formation, overall reactant used, intermediate formation
Step 4. The FAD and dioxygen radical species undergo a colligation reaction to form the FAD-peroxo adduct.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His72A | hydrogen bond acceptor, hydrogen bond donor |
| Tyr201A | hydrogen bond acceptor, hydrogen bond donor |
| Tyr385A | hydrogen bond acceptor, hydrogen bond donor |
Chemical Components
colligation, radical termination, intermediate formation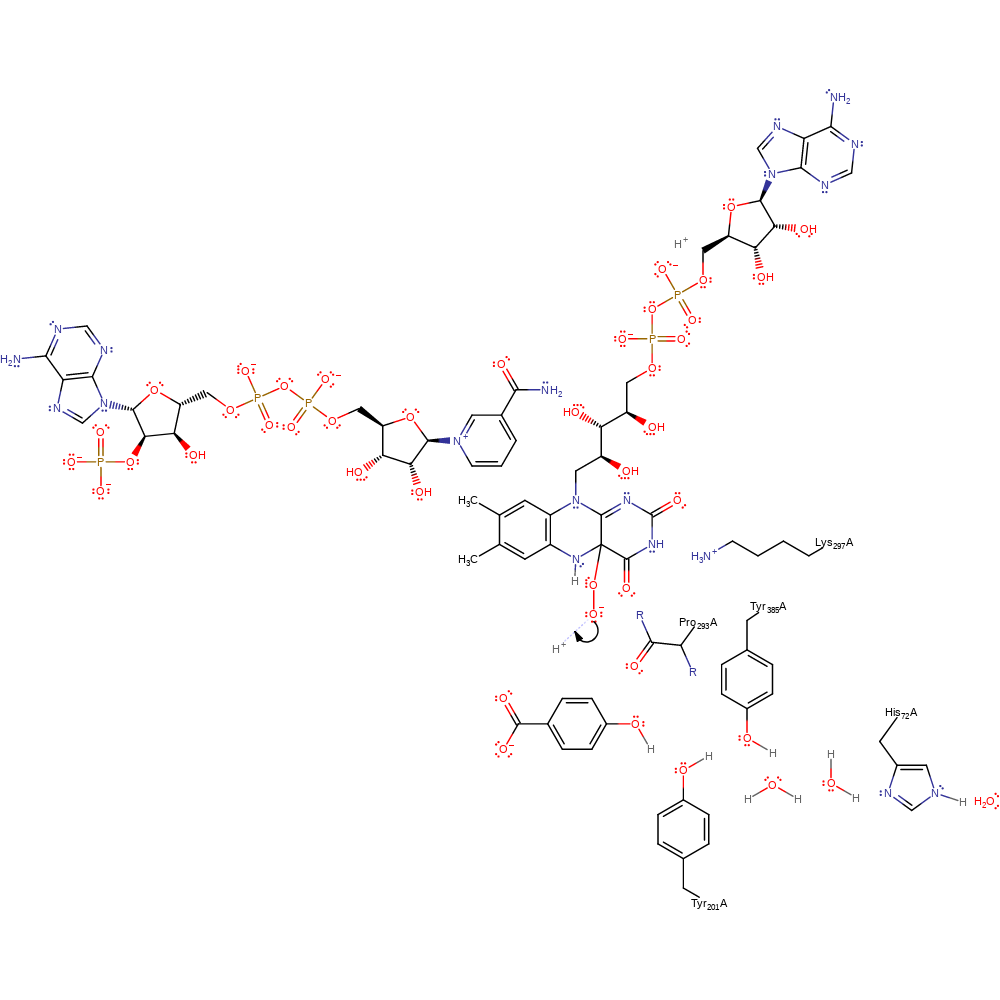
Step 5. Protonation of the flavin peroxide would be favoured by the presence of structural water molecules on the re side of the isoalloxazine that have contact with solvent molecules through a channel into the re side of the flavin proximal to the region of the ribityl side chain [PMID:15581585]. Moreover, the rate of formation of the flavin hydroperoxide is not influenced by pH change, which indicates that the proton required for this reaction does not come from the H-bond network [PMID:15568817].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His72A | hydrogen bond acceptor, hydrogen bond donor |
| Tyr201A | hydrogen bond acceptor, hydrogen bond donor |
| Tyr385A | hydrogen bond acceptor, hydrogen bond donor |
Chemical Components
proton transfer, intermediate formation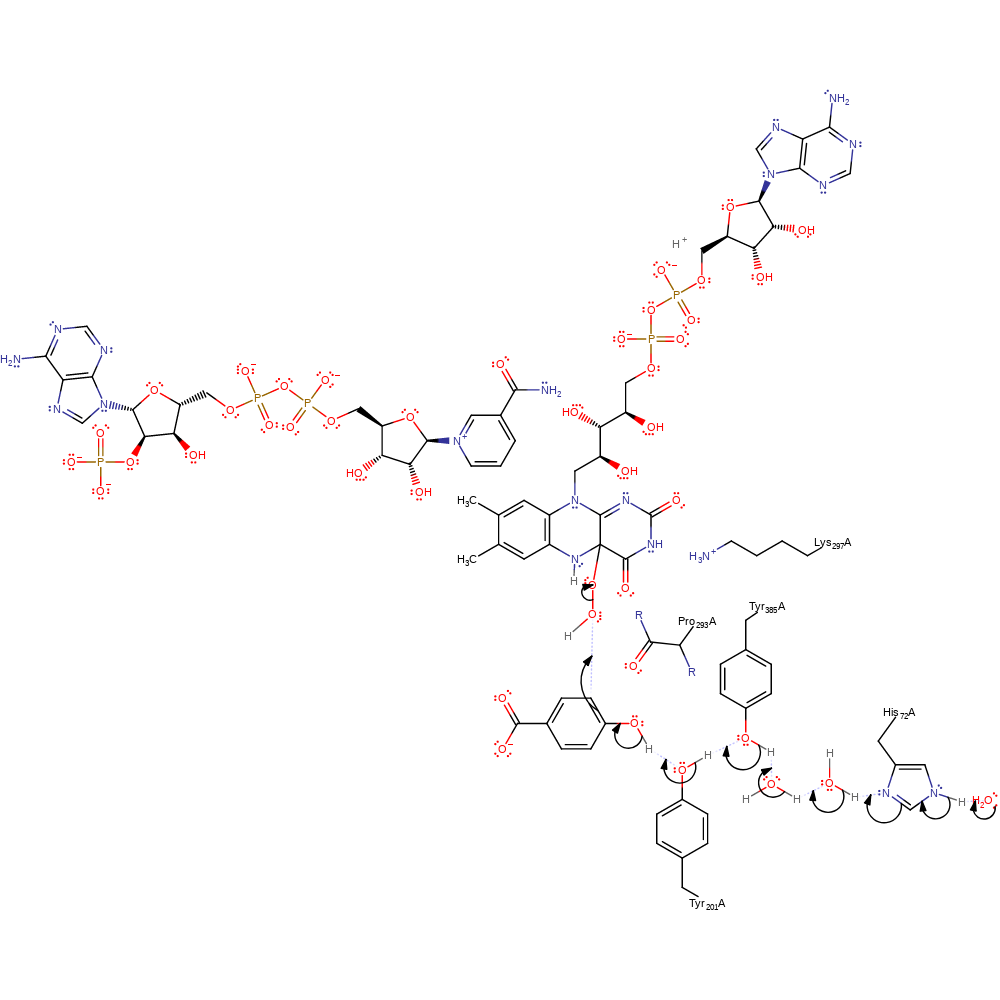
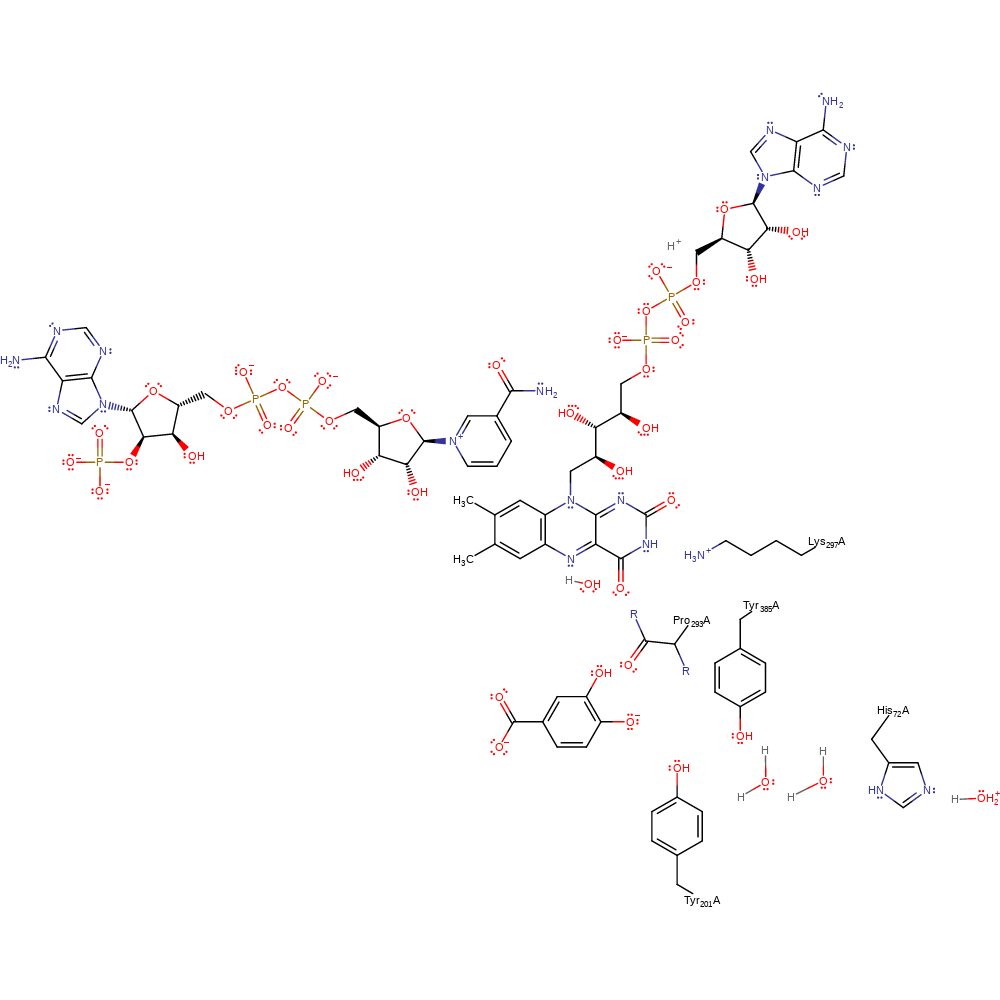
Step 6. In a proton transfer chain involving His72-water-water-Tyr385-Tyr201 water deprotonates the 4-hydroxybenzoate substrate. The substrate undergoes a double bond rearrangement that results in the ortho-position attacking the FAD-peroxo adduct in a nucleophilic substitution that cleaves the O-O bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His72A | hydrogen bond acceptor, hydrogen bond donor, proton relay |
| Tyr201A | hydrogen bond donor, hydrogen bond acceptor, proton relay |
| Tyr385A | hydrogen bond donor, hydrogen bond acceptor, proton relay |
| Lys297A | attractive charge-charge interaction, electrostatic stabiliser |
| Tyr201A | proton donor, proton acceptor |
| Tyr385A | proton donor, proton acceptor |
| His72A | proton acceptor, proton donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, intermediate formation, proton relay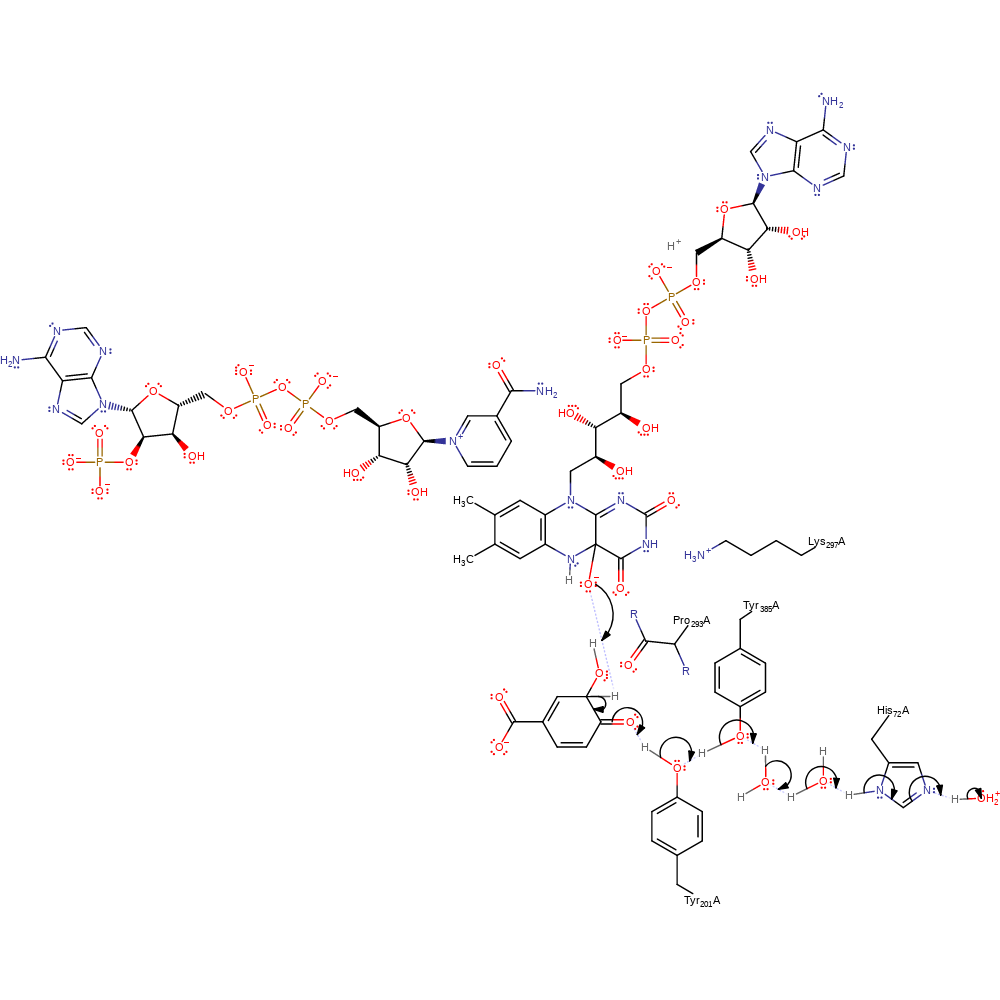
Step 7. The FAD intermediate deprotonates the hydorxylated aromatic intermediate, cleaving the C-H bond and initiating a double bond rearrangement to yield 3,4-dihydroxybenzoic acid. This then takes a proton from water through the His72-water-water-Tyr385-Tyr201 proton transfer chain.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His72A | hydrogen bond acceptor, hydrogen bond donor, proton relay |
| Tyr201A | hydrogen bond donor, hydrogen bond acceptor, proton relay |
| Tyr385A | hydrogen bond donor, hydrogen bond acceptor, proton relay |
| Lys297A | attractive charge-charge interaction, electrostatic stabiliser |
| Tyr201A | proton acceptor |
| Tyr385A | proton acceptor |
| His72A | proton donor, proton acceptor |
| Tyr385A | proton donor |
| Tyr201A | proton donor |
Chemical Components
proton transfer, assisted keto-enol tautomerisation, intermediate formation, proton relay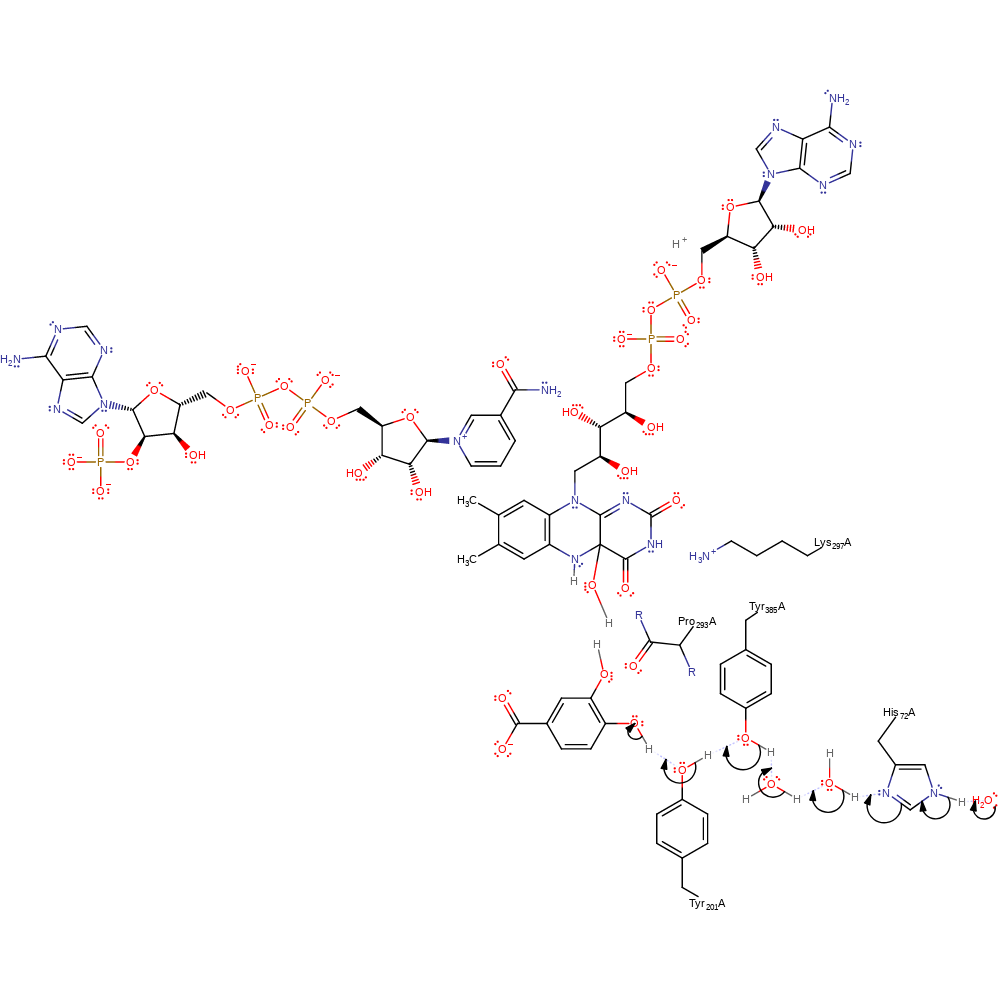
Step 8. Water deprotonates the 3,4-dihydroxybenzoic acid through the His72-water-water-Tyr385-Tyr201 proton transfer chain.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His72A | hydrogen bond acceptor, hydrogen bond donor, proton relay |
| Tyr201A | hydrogen bond donor, hydrogen bond acceptor, proton relay |
| Tyr385A | hydrogen bond donor, hydrogen bond acceptor, proton relay |
| Pro293A (main-C) | hydrogen bond acceptor |
| Lys297A | steric role |
| Pro293A (main-C) | electrostatic stabiliser |
| His72A | proton acceptor |
| Tyr385A | proton acceptor |
| Tyr201A | proton acceptor, proton donor |
| Tyr385A | proton donor |
| His72A | proton donor |
Chemical Components
proton transfer, intermediate terminated, overall product formed, proton relay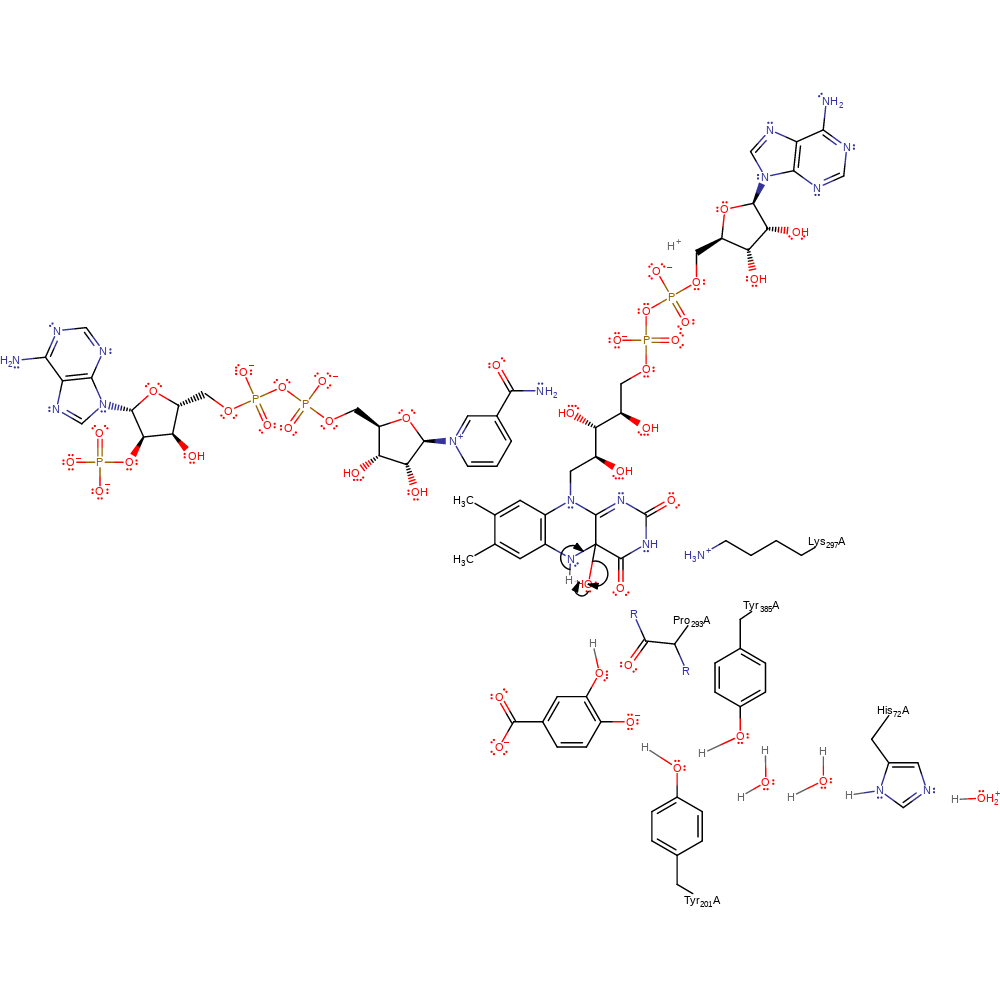
Step 9. The FAD-bound hydroxyl group initiates an intramolecular elimination of water, regenerating the FAD cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His72A | hydrogen bond acceptor, hydrogen bond donor |
| Tyr201A | hydrogen bond acceptor, hydrogen bond donor |
| Tyr385A | hydrogen bond acceptor, hydrogen bond donor |
| Pro293A (main-C) | hydrogen bond acceptor |







 Download:
Download: