Urate oxidase
Urate oxidase is an essential enzyme involved in purine degradation pathway. It is responsible for the conversion of uric acid to allantoin, thus catalysing the oxidative ring opening of the purine ring in the degradative pathway.
The enzyme is lacking in humans, it can therefore be used as a protein drug to treat / reduce toxic uric acid accumulation.
The active enzyme is a homotetramer, each dimer can be considered as a porin-like structure, with the 16 antiparallel strands superimposed in a reverse handed manner on those from a porin, the helices taking the place of the membrane. The enzyme was originally thought to require a copper cofactor, however it has since been shown that is doesn't require a cofactor [PMID:12680763].
The enzyme is a member of a family of proteins with multimeric barrels face to face related by two-fold symmetry, proposed to be a new family of tunnel-shaped proteins.
Reference Protein and Structure
- Sequence
-
Q00511
 (1.7.3.3)
(1.7.3.3)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Aspergillus flavus (Fungus)

- PDB
-
1wrr
- Urate oxidase from aspergillus flavus complexed with 5-amino 6-nitro uracil
(1.64 Å)



- Catalytic CATH Domains
-
3.10.270.10
 (see all for 1wrr)
(see all for 1wrr)
Enzyme Reaction (EC:1.7.3.3)
Enzyme Mechanism
Introduction
Urate oxidase catalyses the first step of the oxidation of uric acid. This reaction is followed by several uncatalysed steps to give allantoin. There is no prosthetic group or metal ion present in the catalytic reaction. Most of the enzyme's catalytic activity results from isolating its substrates in the optimum geometry for the reaction to occur. Uric acid is stabilised as a dianion by urate oxidase through hydrogen bonds with ARG 176 and GLN 228. GLN 228 is positioned to anchor the purine ring.
Catalytic Residues Roles
| UniProt | PDB* (1wrr) | ||
| Lys11, Thr58, His257 | Lys10A, Thr57A, His256A(AA) | These residues form a Thr-Lys-His catalytic triad that is responsible for the transfer of protons to and from the active site. All three residues act as general acid/bases. | proton relay, hydrogen bond donor, proton acceptor, proton donor |
| Arg177, Gln229 | Arg176A(AA), Gln228A(AA) | Help bind and stabilise the substrate in the active site. | electrostatic stabiliser |
Chemical Components
proton transfer, assisted tautomerisation (not keto-enol), overall reactant used, intermediate formation, proton relay, radical formation, electron transfer, colligation, radical termination, intramolecular elimination, intermediate collapse, overall product formed, rate-determining step, bimolecular electrophilic addition, intermediate terminated, native state of enzyme regeneratedReferences
- Imhoff RD et al. (2003), Biochemistry, 42, 4094-4100. General Base Catalysis in the Urate Oxidase Reaction: Evidence for a Novel Thr−Lys Catalytic Diad†. DOI:10.1021/bi027377x. PMID:12680763.
- Colloc'h N et al. (2014), FEBS Lett, 588, 1715-1719. Functional relevance of the internal hydrophobic cavity of urate oxidase. DOI:10.1016/j.febslet.2014.03.017. PMID:24657440.
- Bui S et al. (2014), Angew Chem Int Ed Engl, 53, 13710-13714. Direct Evidence for a Peroxide Intermediate and a Reactive Enzyme-Substrate-Dioxygen Configuration in a Cofactor-free Oxidase. DOI:10.1002/anie.201405485. PMID:25314114.
- Oksanen E et al. (2014), PLoS One, 9, e86651-. The Neutron Structure of Urate Oxidase Resolves a Long-Standing Mechanistic Conundrum and Reveals Unexpected Changes in Protonation. DOI:10.1371/journal.pone.0086651. PMID:24466188.
- Gabison L et al. (2011), Proteins, 79, 1964-1976. X-ray, ESR, and quantum mechanics studies unravel a spin well in the cofactor-less urate oxidase. DOI:10.1002/prot.23022. PMID:21491497.
- Altarsha M et al. (2009), Bioorg Chem, 37, 111-125. Intrinsic reactivity of uric acid with dioxygen: Towards the elucidation of the catalytic mechanism of urate oxidase. DOI:10.1016/j.bioorg.2009.05.004. PMID:19539344.
- Colloc'h N et al. (2008), Biophys J, 95, 2415-2422. Oxygen Pressurized X-Ray Crystallography: Probing the Dioxygen Binding Site in Cofactorless Urate Oxidase and Implications for Its Catalytic Mechanism. DOI:10.1529/biophysj.107.122184. PMID:18375516.
- Gabison L et al. (2008), BMC Struct Biol, 8, 32-. Structural analysis of urate oxidase in complex with its natural substrate inhibited by cyanide: Mechanistic implications. DOI:10.1186/1472-6807-8-32. PMID:18638417.
- Gabison L et al. (2006), FEBS Lett, 580, 2087-2091. Recapture of [S]-allantoin, the product of the two-step degradation of uric acid, by urate oxidase. DOI:10.1016/j.febslet.2006.03.007. PMID:16545381.
- Retailleau P et al. (2004), Acta Crystallogr D Biol Crystallogr, 60, 453-462. Complexed and ligand-free high-resolution structures of urate oxidase (Uox) fromAspergillus flavus: a reassignment of the active-site binding mode. DOI:10.1107/s0907444903029718. PMID:14993669.
- Colloc'h N et al. (1997), Nat Struct Biol, 4, 947-952. Crystal Structure of the protein drug urate oxidase-inhibitor complex at 2.05 Å resolution. DOI:10.1038/nsb1197-947. PMID:9360612.
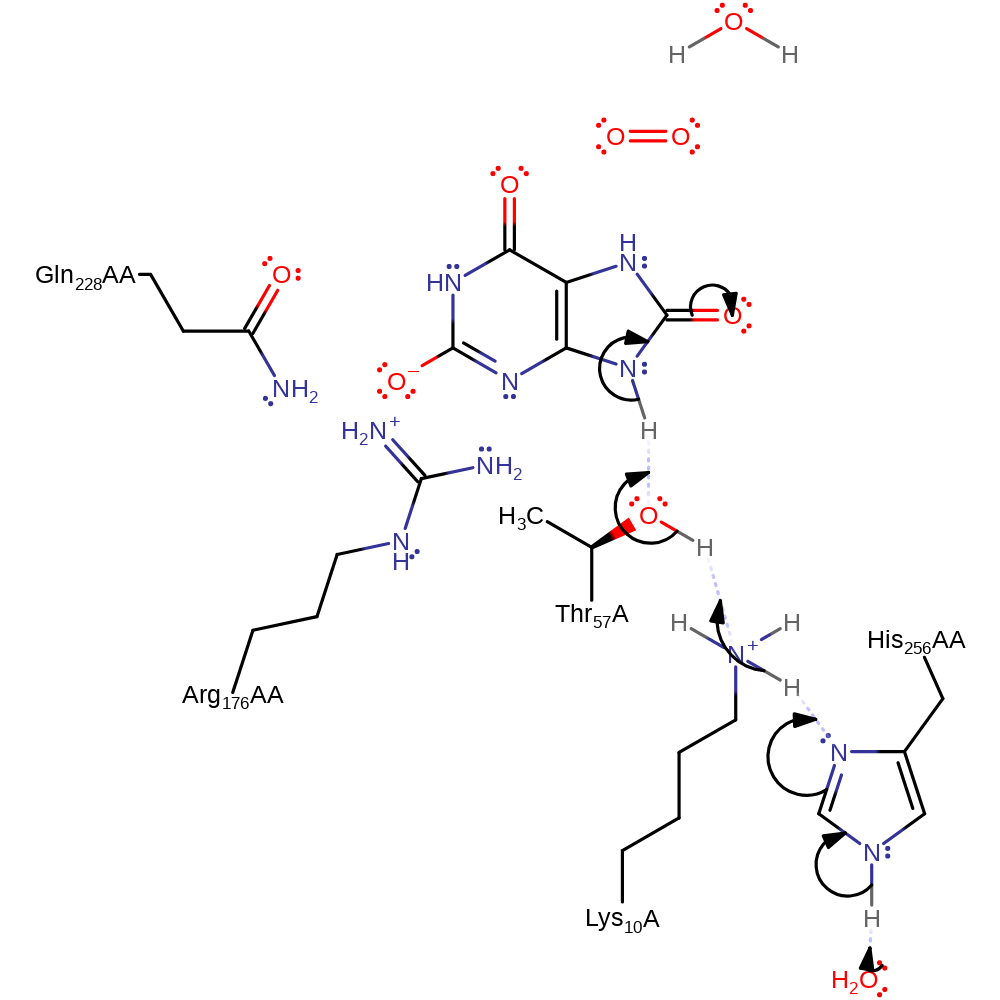
Step 1. Water deprotonates His256AA, which deprotonates the urate substrate through Lys10 and Thr57.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr57A | hydrogen bond acceptor, proton relay |
| Lys10A | hydrogen bond donor, proton relay |
| His256A(AA) | hydrogen bond acceptor, hydrogen bond donor, proton relay |
| Gln228A(AA) | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
| Arg176A(AA) | electrostatic stabiliser |
| Lys10A | proton donor, proton acceptor |
| Thr57A | proton acceptor |
| His256A(AA) | proton donor |
| Thr57A | proton donor |
| His256A(AA) | proton acceptor |
Chemical Components
proton transfer, assisted tautomerisation (not keto-enol), overall reactant used, intermediate formation, proton relay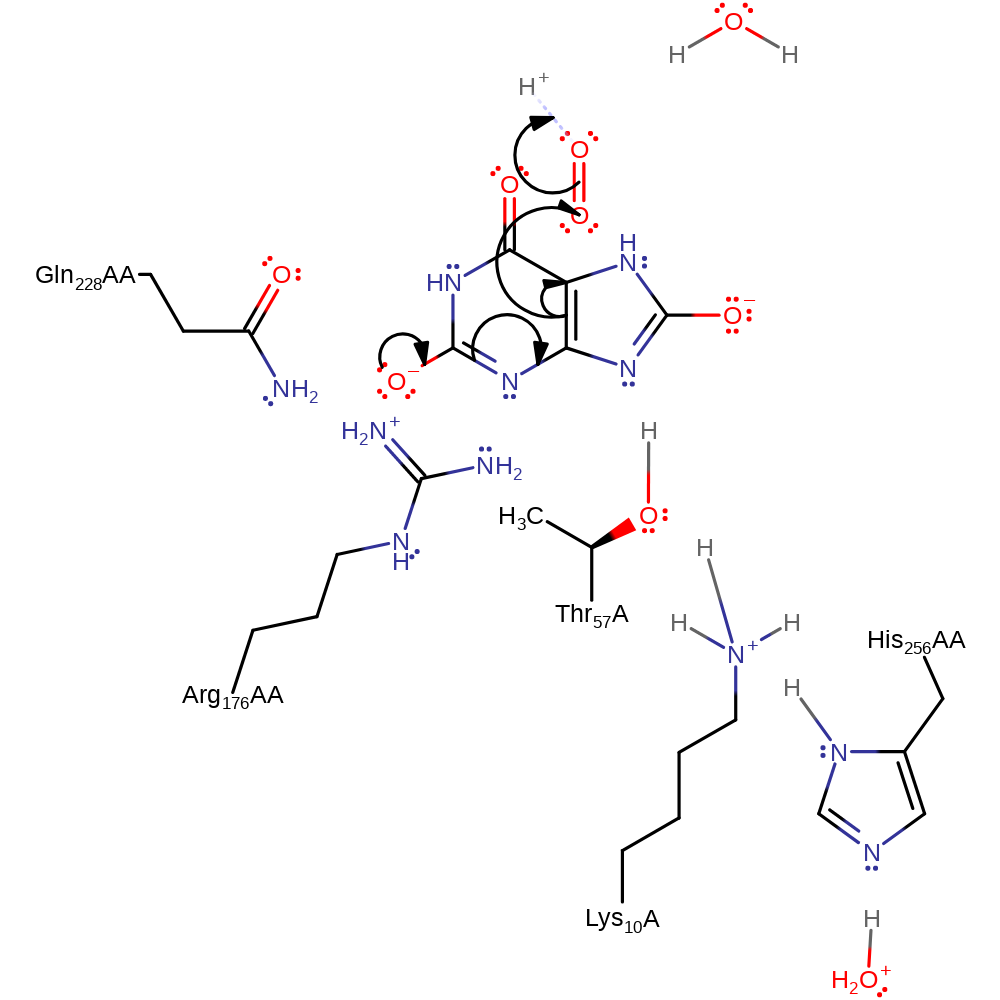
Step 2. The C2 oxyanion collapses initiating a single electron transfer to dioxygen, which deprotonates water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr57A | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
| Lys10A | hydrogen bond donor |
| Gln228A(AA) | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
| Arg176A(AA) | electrostatic stabiliser |
Chemical Components
radical formation, electron transfer, proton transfer, overall reactant used, intermediate formation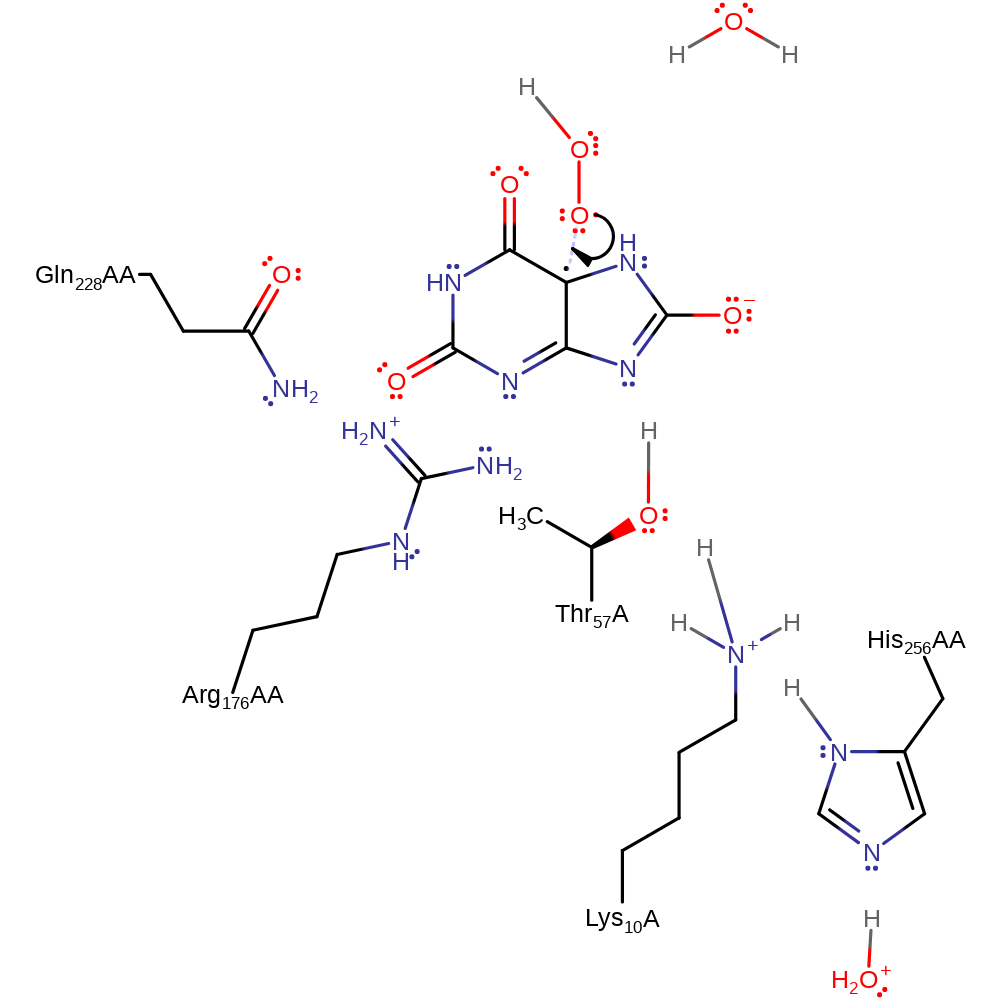
Step 3. The dioxygen radical undergoes a homolytic reaction and colligates to the urate radical.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr57A | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
| Lys10A | hydrogen bond donor |
| Gln228A(AA) | hydrogen bond acceptor, hydrogen bond donor |
| Arg176A(AA) | electrostatic stabiliser |
Chemical Components
colligation, radical termination, intermediate formation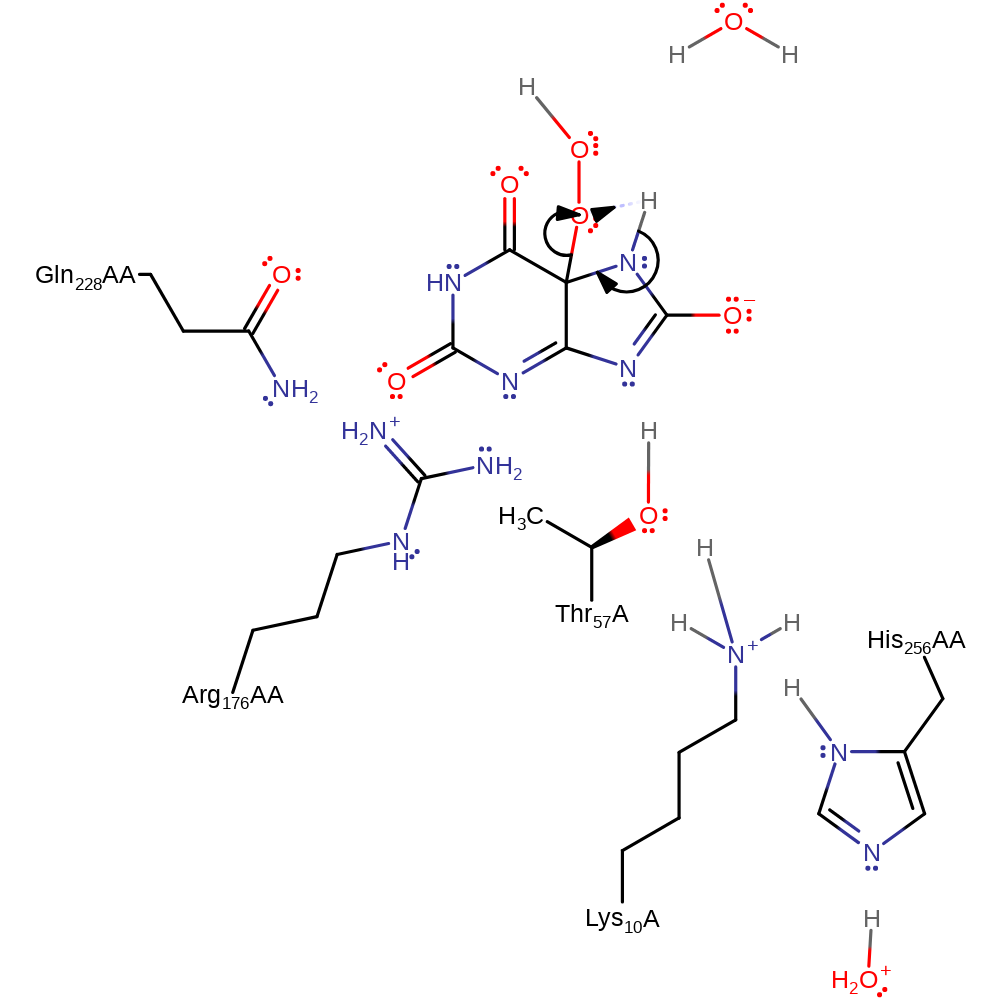
Step 4. The peroxo group deprotonates urate hydroperoxide, which initiates the elimination of hydrogen peroxide.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr57A | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
| Lys10A | hydrogen bond donor |
| Gln228A(AA) | hydrogen bond acceptor, hydrogen bond donor |
| Arg176A(AA) | electrostatic stabiliser |
Chemical Components
ingold: intramolecular elimination, intermediate collapse, intermediate formation, overall product formed, rate-determining step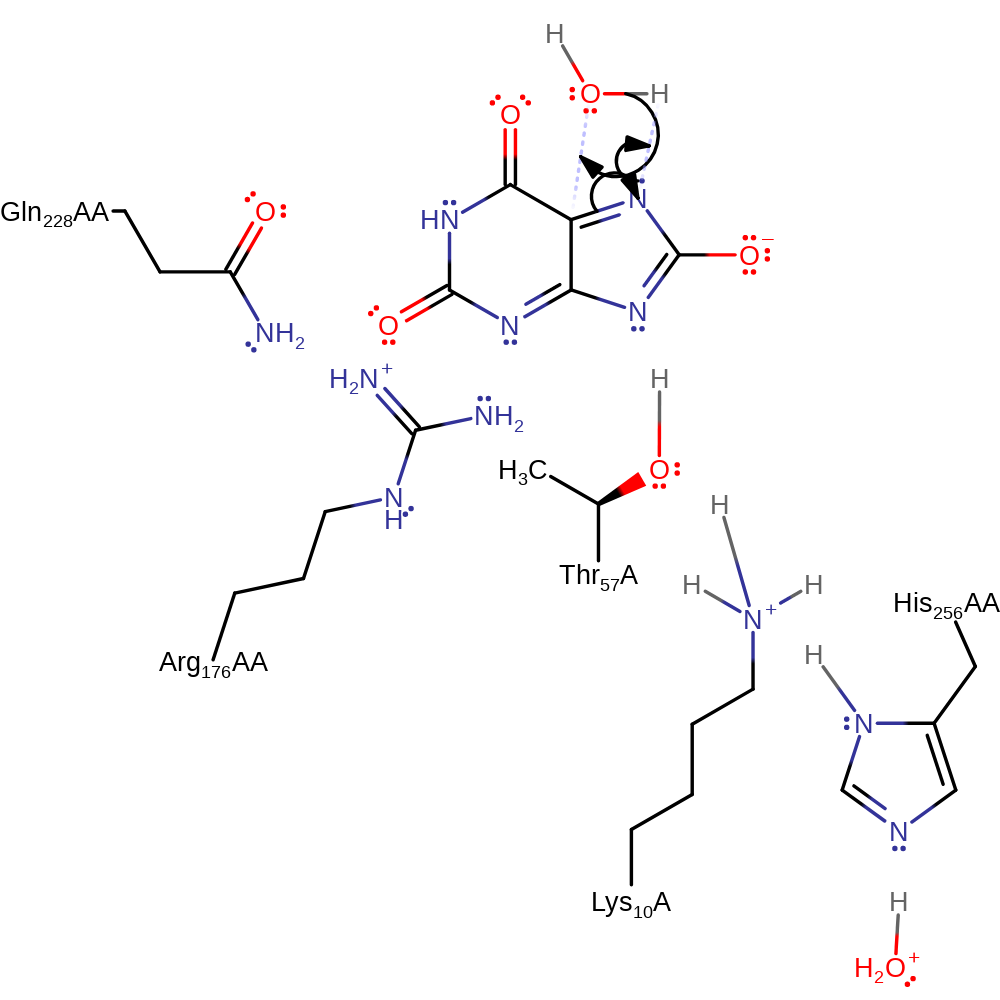
Step 5. N7 of the substrate deprotonates water, which initiates a nucleophilic attack on the C5 of the substrate in an addition reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr57A | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
| Lys10A | hydrogen bond donor |
| Gln228A(AA) | hydrogen bond acceptor, hydrogen bond donor |
| Arg176A(AA) | electrostatic stabiliser |





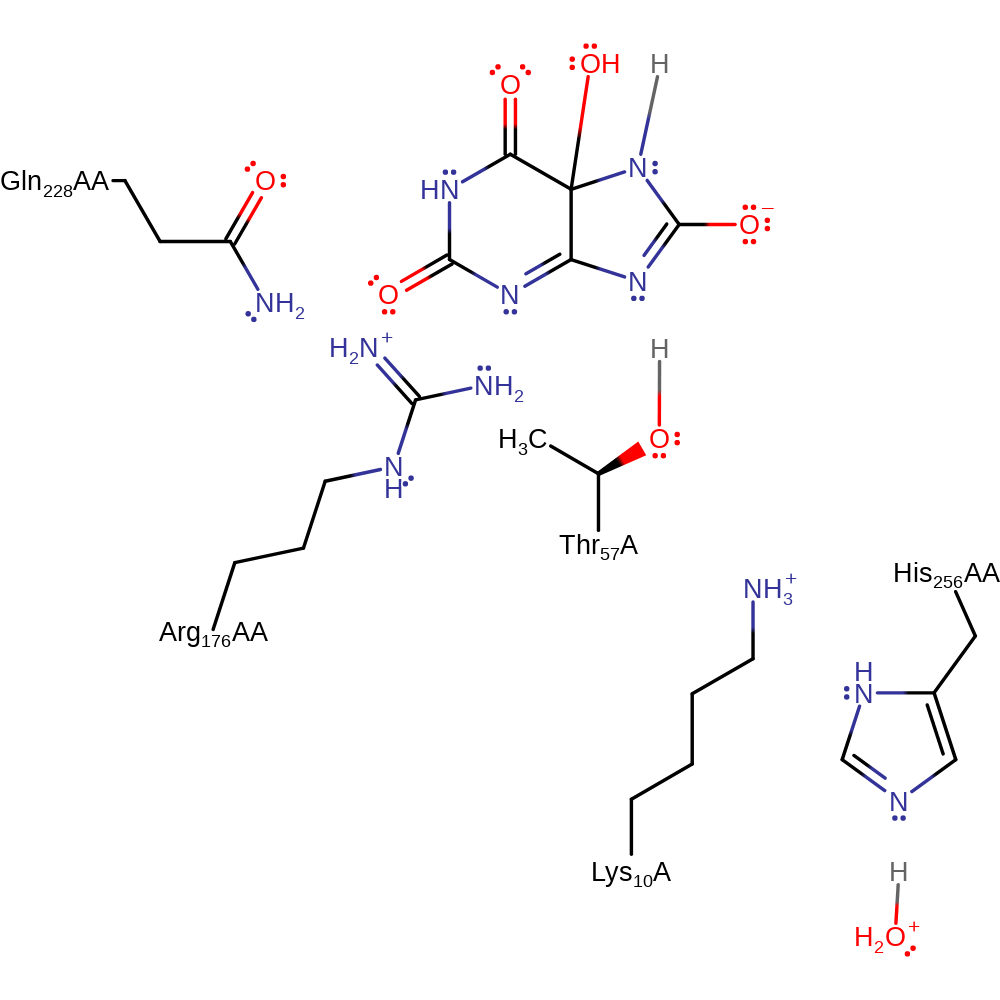 Download:
Download: