NAD(P)+ transhydrogenase (AB-specific)
Transhydrogenase, isolated from Rhodospirillum rubrum, is a transmembrane protein that catalyses the hydride transfer from NADH to NADP+. The driving force for this reaction is the movement of a proton down a proton electrochemical gradient, with one proton transfer per hydride transfer, from the periplasm to the cytoplasm. Domain I (located in the PDB file) binds NADH and contains the catalytic residues for the reaction. Domain II facilitates proton translocation and domain III binds NADP+. Proton transfer through domain II propagates a conformational change from domain II through domain III to domain I. These conformational changes are required to align the substrates correctly and so couples proton translocation to hydride transfer.
Reference Protein and Structure
- Sequences
-
Q2RSB2
 (7.1.1.1)
(7.1.1.1)
Q2RSB4 (7.1.1.1)
(7.1.1.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Rhodospirillum rubrum ATCC 11170 (Bacteria)

- PDB
-
1hzz
- THE ASYMMETRIC COMPLEX OF THE TWO NUCLEOTIDE-BINDING COMPONENTS (DI, DIII) OF PROTON-TRANSLOCATING TRANSHYDROGENASE
(2.5 Å)



- Catalytic CATH Domains
-
3.40.50.1220
 3.40.50.720
3.40.50.720  (see all for 1hzz)
(see all for 1hzz)
Enzyme Reaction (EC:1.6.1.2)
Enzyme Mechanism
Introduction
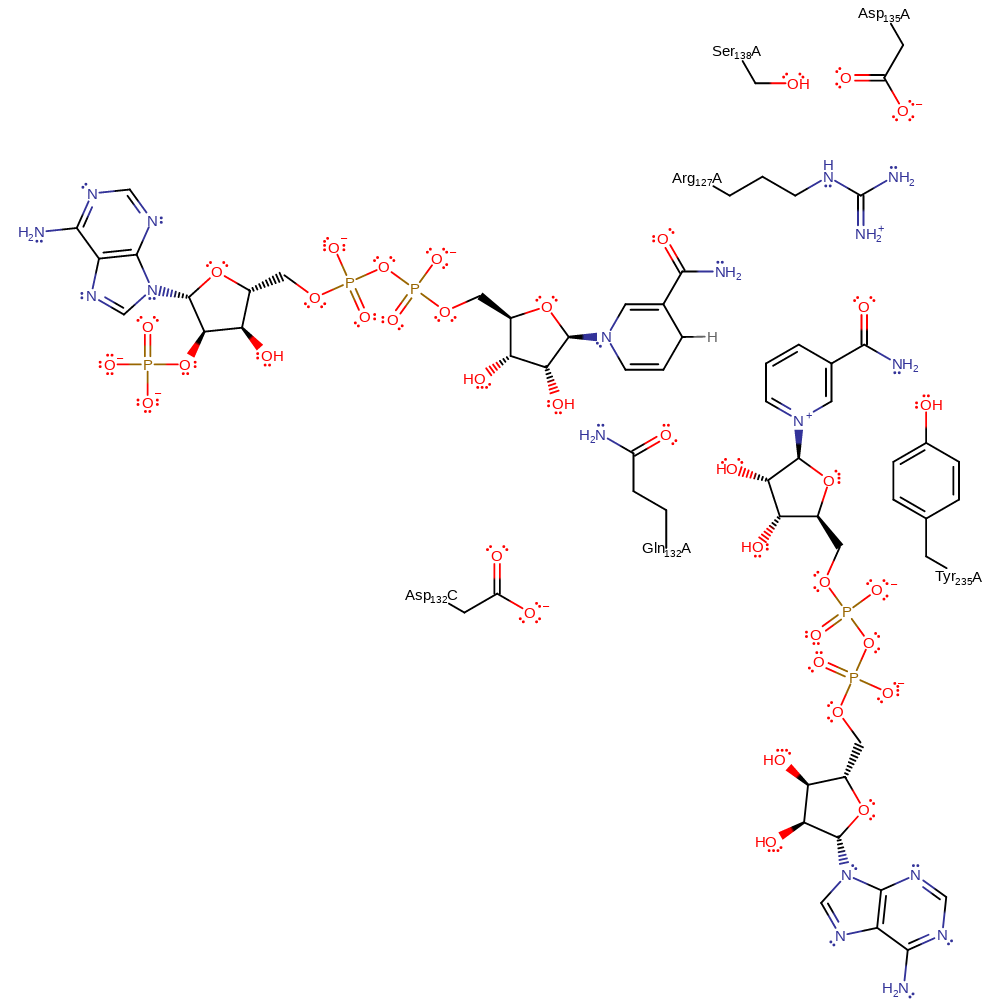
The translocation of the proton from outside the membrane to the inside causes a conformational rearrangement within all domains such that Arg127 of domain I is positioned to stabilise NADH in the proximal position. This allows NADH and NADP+ to align correctly and hydride transfer occurs between the C4 positions of the nicotinic rings of both substrates. The NADP+ and NADH molecules are tethered in place by Gln132. The conformational change causes Arg127 to interact with Asp135 and Tyr235, which in turn causes the Tyr235 to close off the active site.
Catalytic Residues Roles
| UniProt | PDB* (1hzz) | ||
| Tyr235 | Tyr235B | Prevents competing reactions from occuring by blocking solvent access to the active site. | steric role, polar/non-polar interaction |
| Ser138 | Ser138B | Ser138 forms a hydrogen bond to Asp135, which strengthens the hydrogen bond between Asp135 and Arg127. This ensures that NADH is maintained in the proximal position. | electrostatic stabiliser |
| Asp393 | Asp132C | Asp132 is the acceptor of the proton which comes from outside the membrane. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Arg127 | Arg127B | Arg127 is positioned to stabilise the proximal position of NADH and allow hydride transfer. Arg127 is held in this position by a hydrogen bond to Asp135. | hydrogen bond donor, steric role |
| Asp135 | Asp135B | Asp135 forms a hydrogen bond to Arg127 and stabilises the position of the latter to ensure the correct positioning of NADH. The hydrogen bond between the two residues is strengthened by hydrogen bonds between Asp135 and Ser138, and Asp135 and Gln132. | hydrogen bond acceptor, steric role |
| Gln132 | Gln132B | Gln132 forms a hydrogen bond to Asp135, which strengthens the hydrogen bond between Asp135 and Arg127. This ensures that NADH is maintained in the proximal position. | steric locator |
Chemical Components
proton transfer, aromatic unimolecular elimination by the conjugate base, hydride transfer, aromatic bimolecular nucleophilic addition, overall reactant used, overall product formed, native state of enzyme regeneratedReferences
- Brondijk TH et al. (2006), J Biol Chem, 281, 13345-13354. The Role of Invariant Amino Acid Residues at the Hydride Transfer Site of Proton-translocating Transhydrogenase. DOI:10.1074/jbc.m513230200. PMID:16533815.
- Pedersen A et al. (2008), J Bioenerg Biomembr, 40, 463-473. Proton-translocating transhydrogenase: an update of unsolved and controversial issues. DOI:10.1007/s10863-008-9170-x. PMID:18972197.
- Obiozo UM et al. (2007), J Biol Chem, 282, 36434-36443. Substitution of Tyrosine 146 in the dI Component of Proton-translocating Transhydrogenase Leads to Reversible Dissociation of the Active Dimer into Inactive Monomers. DOI:10.1074/jbc.m705433200. PMID:17911104.
- O'Connell KM et al. (2004), Biochemistry, 43, 10965-10978. Differential Inhibition of Six Copper Amine Oxidases by a Family of 4-(Aryloxy)-2-butynamines: Evidence for a New Mode of Inactivation†. DOI:10.1021/bi0492004. PMID:15323556.
- van Boxel GI et al. (2003), Biochemistry, 42, 1217-1226. Glutamine 132 in the NAD(H)-Binding Component of Proton-Translocating Transhydrogenase Tethers the Nucleotides before Hydride Transfer†. DOI:10.1021/bi027032e. PMID:12564924.
- Singh A et al. (2003), J Biol Chem, 278, 33208-33216. Interactions between Transhydrogenase and Thio-nicotinamide Analogues of NAD(H) and NADP(H) Underline the Importance of Nucleotide Conformational Changes in Coupling to Proton Translocation. DOI:10.1074/jbc.m303061200. PMID:12791694.
- Rodrigues DJ et al. (2001), Eur J Biochem, 268, 1430-1438. A change in ionization of the NADP(H)-binding component (dIII) of proton-translocating transhydrogenase regulates both hydride transfer and nucleotide release. DOI:10.1046/j.1432-1327.2001.02008.x. PMID:11231296.
- Cotton NP et al. (2001), Structure, 9, 165-176. The crystal structure of an asymmetric complex of the two nucleotide binding components of proton-translocating transhydrogenase. PMID:11250201.

Step 1. Asp132C obtains a proton, causing a rearrangement of the active site. The full proton translocation pathways is not well defined. However, as a consequence of a step in the proton translocation pathway a change in the ionisation of a group (possibly Asp132C) leads to a transition between the open and occluded states. This transition brings the C4 atoms of the NADP+ and NADH molecules into a position where the hydride transfer can occur. The transition also causes the Arg127 to become deeply buried within the active site and to form H bonds with Asp135, thus pulling in the side chain of Tyr235 to completely seal the site. This prevents solvent from accessing the active site and thus avoids undesired reactions.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp132C | hydrogen bond acceptor |
| Gln132B | steric locator |
| Ser138B | electrostatic stabiliser |
| Asp132C | proton acceptor |
Chemical Components
proton transferCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg127B | hydrogen bond donor, steric role |
| Asp135B | hydrogen bond acceptor, steric role |
| Tyr235B | steric role, polar/non-polar interaction |
| Gln132B | steric locator |
| Ser138B | electrostatic stabiliser |
Chemical Components
ingold: aromatic unimolecular elimination by the conjugate base, hydride transfer, ingold: aromatic bimolecular nucleophilic addition, overall reactant used, overall product formed
Step 3. Asp132C releases its proton. The release of the products causes the proton to be released to the other side of the membrane and therefore the active site to return to the open state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg127B | hydrogen bond donor, steric role |
| Asp135B | hydrogen bond acceptor, steric role |
| Asp132C | hydrogen bond donor |
| Ser138B | electrostatic stabiliser |
| Gln132B | steric locator |
| Asp132C | proton donor |





 Download:
Download: