Sarcosine oxidase
Monomeric sarcosine oxidase (MSOX) catalyses the oxidative demethylation of sarcosine (N-methylglycine) to form glycine and formaldehyde. Sarcosine is a common soil metabolite that can act as the sole source of carbon and energy for many microorganisms capable of expressing sarcosine oxidase.
MSOX is part of a family of enzymes which contain flavin (FAD covalently attached to the protein via Cys315) and catalyse the oxidation of various secondary and tertiary amino acids.
Despite the vast amount of research that has been done on these enzymes, the exact mechanism remains elusive. There are three proposals: polar, hydride and single electron transfer. To date, there is little evidence to suggest one over the other.
The mechanism by which Cys315 becomes attached to FAD involves His45 and Arg49 [PMID:10368302] and is thought to be autocatalytic. The formation of the covalent FAD adduct is not shown in this entry.
Reference Protein and Structure
- Sequence
-
P40859
 (1.5.3.1)
(1.5.3.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Bacillus sp. B-0618 (Bacteria)

- PDB
-
2gb0
- Monomeric sarcosine oxidase: structure of a covalently flavinylated amine oxidizing enzyme
(1.85 Å)



- Catalytic CATH Domains
-
3.30.9.10
 3.50.50.60
3.50.50.60  (see all for 2gb0)
(see all for 2gb0)
- Cofactors
- Fadh2(2-) (1), Water (4)
Enzyme Mechanism
Introduction
The polar mechanism involves the formation of a covalent flavin-substrate intermediate in a reversible reaction involving nucleophilic addition of the substrate amino group at flavin C(4a).
Catalytic Residues Roles
| UniProt | PDB* (2gb0) | ||
| Lys266 | Lys265B | Lys265 is hydrogen bonded to the N(5) position of the flavin ring via a bridging water (wat1) and is also hydrogen bonded to a second nearby water (wat2). The identification of Lys265 as the site of oxygen activation strongly suggests that the solvent molecules might define a pre-organised binding site for the superoxide anion that could accelerate the 1-electron reduction of oxygen by lowering the reorganisation energy associated with transforming the surrounding medium. | proton relay, hydrogen bond donor, proton acceptor, proton donor |
| His270 | His269B | His269 appears to be important in optimising the orientation of bound substrate with respect to electron transfer to flavin. It is also postulated to act as a general acid/base in the polar mechanism. However, mutation of this residue suggests that this second function is less likely. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Thr49 | Thr48B | Forms part of a proton relay chain with Lys265 and four water molecules. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, proton relay |
| Cys316 | Cys315B | This residue is covalently attached to the FAD cofactor. This covalent attachment is essential to the enzyme's activity and is thought to function by modulating the redox potential of the FAD and holding the weakly bound FAD in the active site. | covalently attached, activator, alter redox potential |
| Arg50 | Arg49B | The side chain of Arg49 is in van der Waals contact with the si-face of the flavin ring and is essential for covalent flavin attachment. It also plays an important role in sarcosine oxidation by virtue of its electrostatic effect on the active site environment. The positively charged guanidinium side chain in contact with the flavin ring raises the flavin reduction potential and thereby facilitates sarcosine oxidation. An additional role for Arg49 in sarcosine oxidation is suggested by the fact that MSOX is known to bind the unreactive zwitterionic form of its amino acid substrates. Substrate activation is achieved by inducing a large decrease in the pKa of the bound zwitterion. Electrostatic interaction of the reactive substrate anion with the positively charged side chain of Arg49 may contribute to the observed shift in the pKa of the bound amino acid. | modifies pKa, electrostatic stabiliser |
| Lys349, His46 | Lys348B, His45B | Help activate and stabilise the flavin cofactor. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
aromatic bimolecular nucleophilic addition, proton transfer, overall reactant used, cofactor used, enzyme-substrate complex formation, intermediate formation, proton relay, bimolecular elimination, enzyme-substrate complex cleavage, intermediate collapse, overall product formed, electron transfer, radical formation, inferred reaction step, bimolecular homolytic addition, aromatic intramolecular elimination, intermediate terminated, native state of cofactor regenerated, native state of enzyme regenerated, reaction occurs outside the enzyme, bimolecular nucleophilic addition, intramolecular eliminationReferences
- Trickey P et al. (1999), Structure, 7, 331-345. Monomeric sarcosine oxidase: structure of a covalently flavinylated amine oxidizing enzyme. DOI:10.2210/pdb1l9f/pdb. PMID:10368302.
- Bucci A et al. (2016), J Chem Theory Comput, 12, 2964-2972. Kinetics of O2Entry and Exit in Monomeric Sarcosine Oxidase via Markovian Milestoning Molecular Dynamics. DOI:10.1021/acs.jctc.6b00071. PMID:27168219.
- Pietra F (2015), Chem Biodivers, 12, 1163-1171. On the Quest of Dioxygen by Monomeric Sarcosine Oxidase. A Molecular Dynamics Investigation. DOI:10.1002/cbdv.201400362. PMID:26265568.
- Bucci A et al. (2014), J Chem Theory Comput, 10, 2668-2676. Oxygen Pathways and Allostery in Monomeric Sarcosine Oxidase via Single-Sweep Free-Energy Reconstruction. DOI:10.1021/ct500088z. PMID:25061440.
- Jorns MS et al. (2010), Biochemistry, 49, 3631-3639. Structural Characterization of Mutations at the Oxygen Activation Site in Monomeric Sarcosine Oxidase,. DOI:10.1021/bi100160j. PMID:20353187.
- Hassan-Abdallah A et al. (2008), Biochemistry, 47, 2913-2922. Arginine 49 Is a Bifunctional Residue Important in Catalysis and Biosynthesis of Monomeric Sarcosine Oxidase: A Context-Sensitive Model for the Electrostatic Impact of Arginine to Lysine Mutations†,‡. DOI:10.1021/bi702351v. PMID:18251505.
- Zhao G et al. (2008), Biochemistry, 47, 9124-9135. Identification of the Oxygen Activation Site in Monomeric Sarcosine Oxidase: Role of Lys265 in Catalysis†. DOI:10.1021/bi8008642. PMID:18693755.
- Hassan-Abdallah A et al. (2008), Biochemistry, 47, 1136-1143. Covalent Flavinylation of Monomeric Sarcosine Oxidase: Identification of a Residue Essential for Holoenzyme Biosynthesis†. DOI:10.1021/bi702077q. PMID:18179257.
- Hassan-Abdallah A et al. (2006), Biochemistry, 45, 9454-9462. Role of the Covalent Flavin Linkage in Monomeric Sarcosine Oxidase†. DOI:10.1021/bi0607352. PMID:16878980.
- Zhao G et al. (2006), Biochemistry, 45, 5985-5992. Spectral and Kinetic Characterization of the Michaelis Charge Transfer Complex in Monomeric Sarcosine Oxidase†. DOI:10.1021/bi0600852. PMID:16681370.
- Zhao G et al. (2005), Biochemistry, 44, 16866-16874. Ionization of Zwitterionic Amine Substrates Bound to Monomeric Sarcosine Oxidase†. DOI:10.1021/bi051898d. PMID:16363800.
- Hassan-Abdallah A et al. (2005), Biochemistry, 44, 6452-6462. Biosynthesis of Covalently Bound Flavin: Isolation and in Vitro Flavinylation of the Monomeric Sarcosine Oxidase Apoprotein†. DOI:10.1021/bi047271x. PMID:15850379.
- Khanna P et al. (2003), Biochemistry, 42, 864-869. Tautomeric Rearrangement of a Dihydroflavin Bound to Monomeric Sarcosine Oxidase orN-Methyltryptophan Oxidase†. DOI:10.1021/bi0206098. PMID:12549903.
- Zhao G et al. (2002), Biochemistry, 41, 9747-9750. Monomeric Sarcosine Oxidase: Evidence for an Ionizable Group in the E·S Complex†. DOI:10.1021/bi020285n.
- Zhao G et al. (2002), Biochemistry, 41, 9751-9764. Monomeric Sarcosine Oxidase: Role of Histidine 269 in Catalysis†,‡. DOI:10.1021/bi020286f.
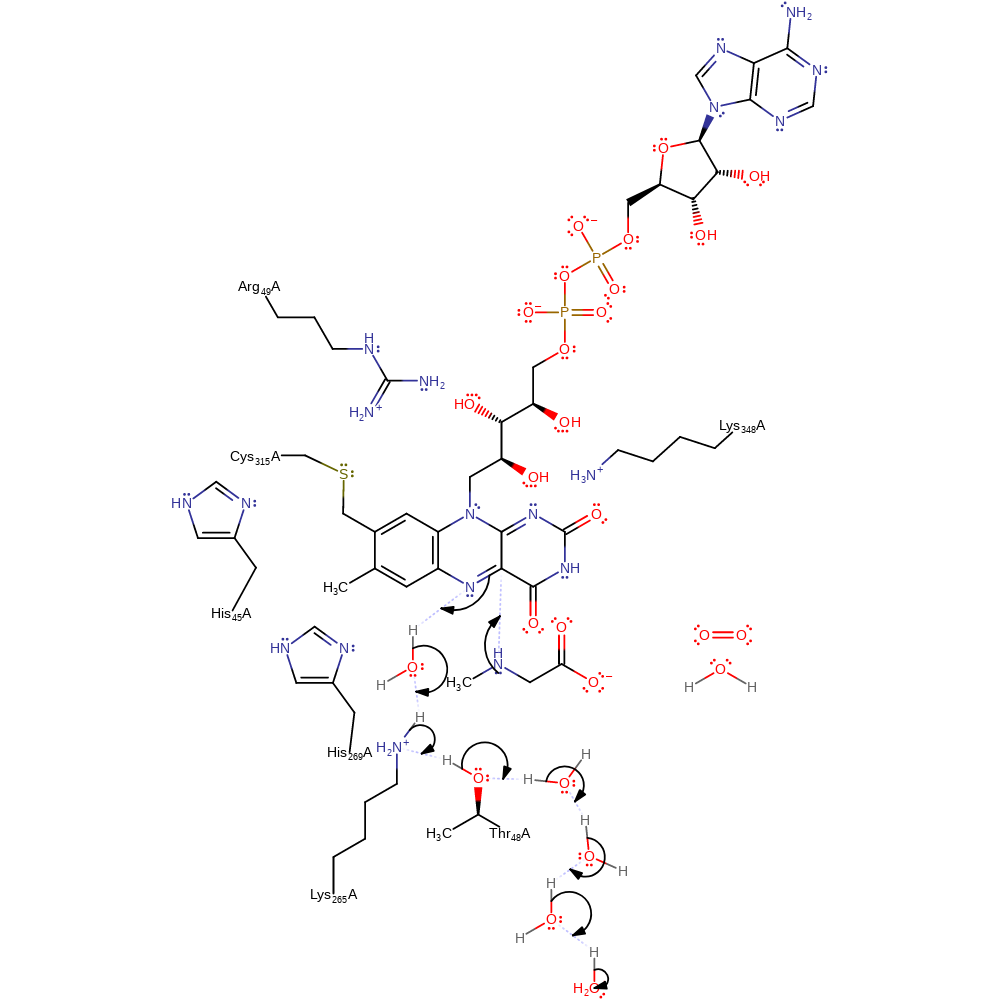
Step 1. The secondary amine of sarcosine initiates a nucleophilic attack on the C4a of FAD in an addition reaction. N5 deprotonates water, which deprotonates Thr48 that regains its proton from bulk solvent through a chain of water molecules.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys315B | alter redox potential |
| Arg49B | modifies pKa |
| His45B | electrostatic stabiliser |
| Lys265B | hydrogen bond donor, proton relay |
| Thr48B | hydrogen bond acceptor, proton relay |
| Cys315B | covalently attached |
| Lys348B | hydrogen bond donor |
| Lys265B | proton donor |
| Thr48B | proton donor |
| Lys265B | proton acceptor |
| Thr48B | proton acceptor |
Chemical Components
ingold: aromatic bimolecular nucleophilic addition, proton transfer, overall reactant used, cofactor used, enzyme-substrate complex formation, intermediate formation, proton relay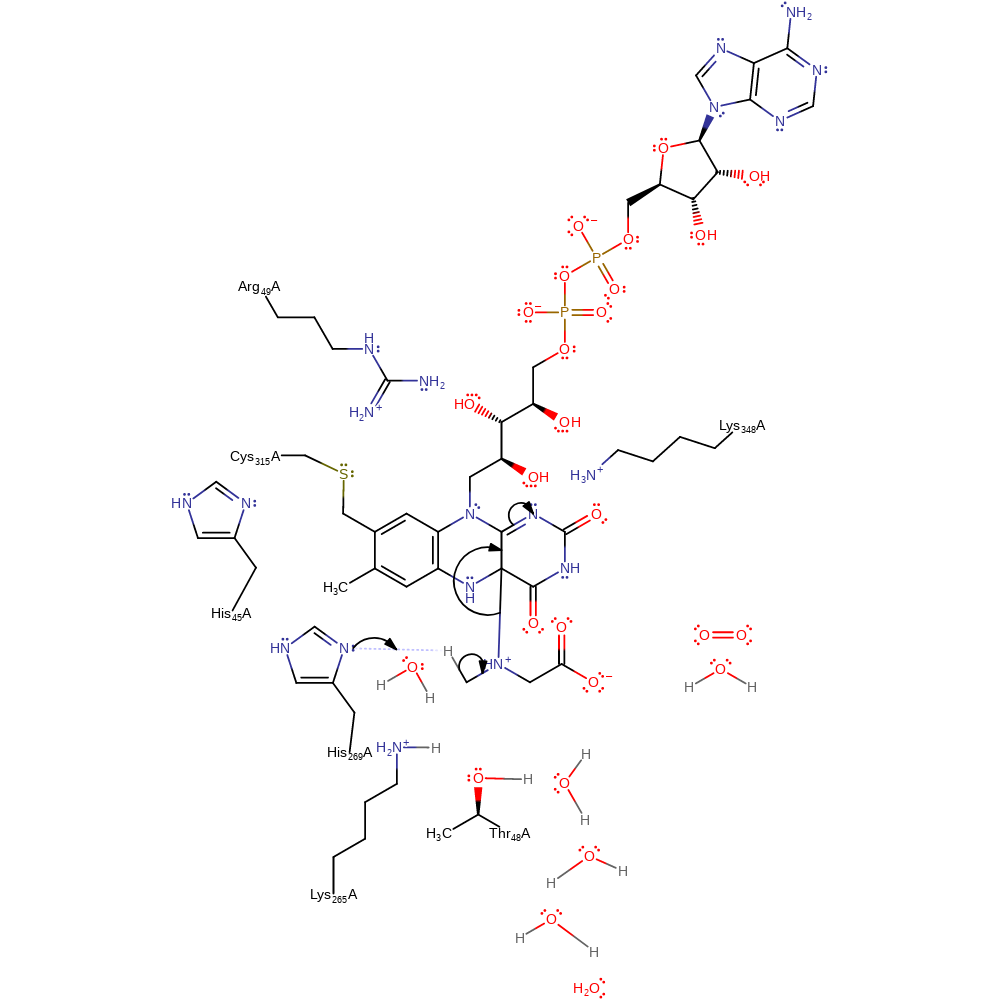
Step 2. His269 deprotonates the terminal methyl group of the FAD-bound sarcosine, eliminating reduced FAD.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His45B | electrostatic stabiliser |
| Cys315B | alter redox potential |
| His269B | hydrogen bond acceptor |
| Lys265B | hydrogen bond donor |
| Thr48B | hydrogen bond acceptor, hydrogen bond donor |
| Cys315B | covalently attached |
| Lys348B | hydrogen bond donor, electrostatic stabiliser |
| His269B | proton acceptor |
Chemical Components
ingold: bimolecular elimination, enzyme-substrate complex cleavage, intermediate collapse, intermediate formation, overall product formed
Step 3. The FAD undergoes double bond rearrangement which causes a single electron to be transferred to a dioxygen molecule.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His269B | hydrogen bond donor |
| Lys265B | hydrogen bond donor |
| Thr48B | hydrogen bond acceptor, hydrogen bond donor |
| Cys315B | covalently attached, activator |
| Lys348B | hydrogen bond donor, electrostatic stabiliser |
| Cys315B | alter redox potential |
| His45B | electrostatic stabiliser |
| Arg49B | modifies pKa |
Chemical Components
electron transfer, radical formation, overall reactant used, intermediate formation, inferred reaction step
Step 4. The dioxygen molecule undergoes a homolytic reaction in which it colligates to FAD, with concomitant deprotonation of His269.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His269B | hydrogen bond donor |
| Lys265B | hydrogen bond donor |
| Thr48B | hydrogen bond acceptor, hydrogen bond donor |
| Cys315B | covalently attached, activator |
| Lys348B | hydrogen bond donor |
| Cys315B | alter redox potential |
| His45B | electrostatic stabiliser |
| Arg49B | electrostatic stabiliser |
| His269B | proton donor |
Chemical Components
ingold: bimolecular homolytic addition, proton transfer, enzyme-substrate complex formation, intermediate formation, inferred reaction step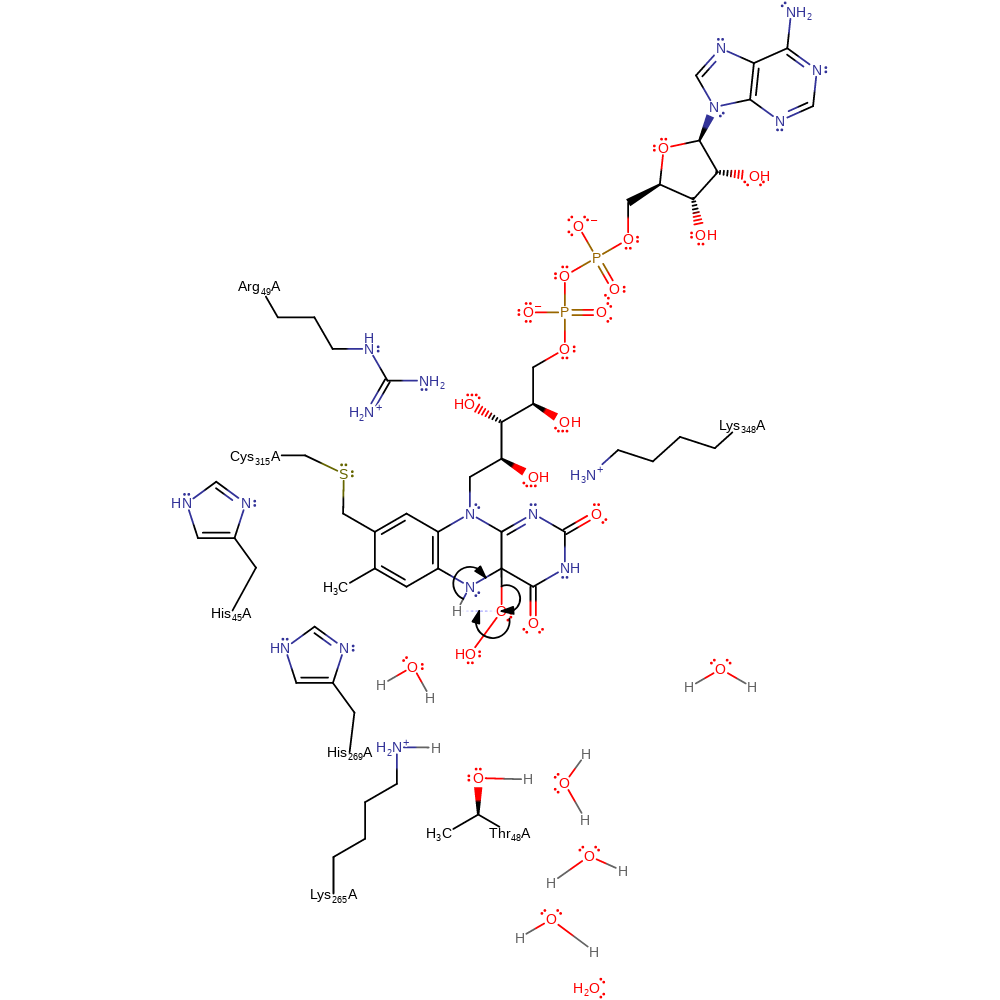
Step 5. The peroxo group deprotonates FAD, which initiates the elimination of hydrogen peroxide.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys265B | hydrogen bond donor |
| Thr48B | hydrogen bond acceptor, hydrogen bond donor |
| Cys315B | covalently attached, activator |
| Lys348B | hydrogen bond donor |
| His45B | electrostatic stabiliser |
| Arg49B | electrostatic stabiliser |
Chemical Components
ingold: aromatic intramolecular elimination, enzyme-substrate complex cleavage, intermediate collapse, intermediate terminated, overall product formed, native state of cofactor regenerated, native state of enzyme regenerated, inferred reaction step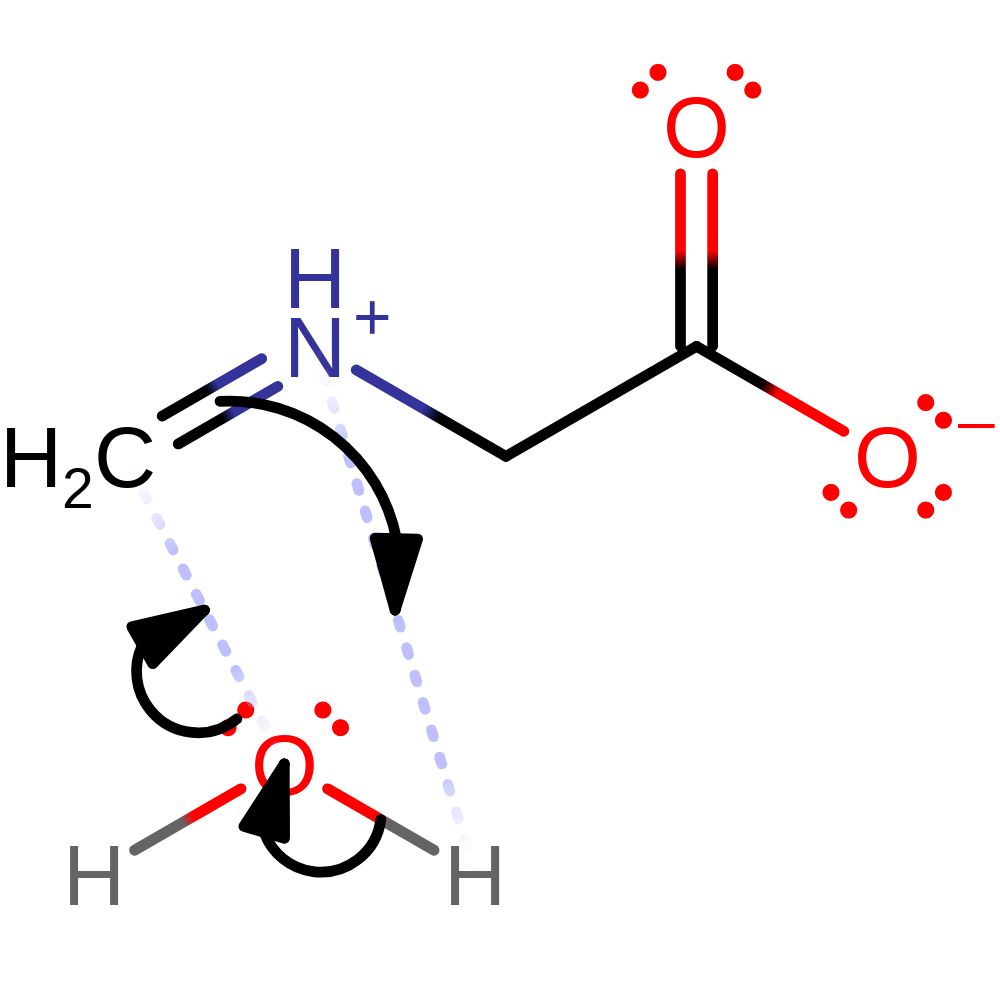
Step 6. The product of the enzyme undergoes spontaneous hydrolysis outside of the active site to produce formaldehyde and glycine.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
reaction occurs outside the enzyme, ingold: bimolecular nucleophilic addition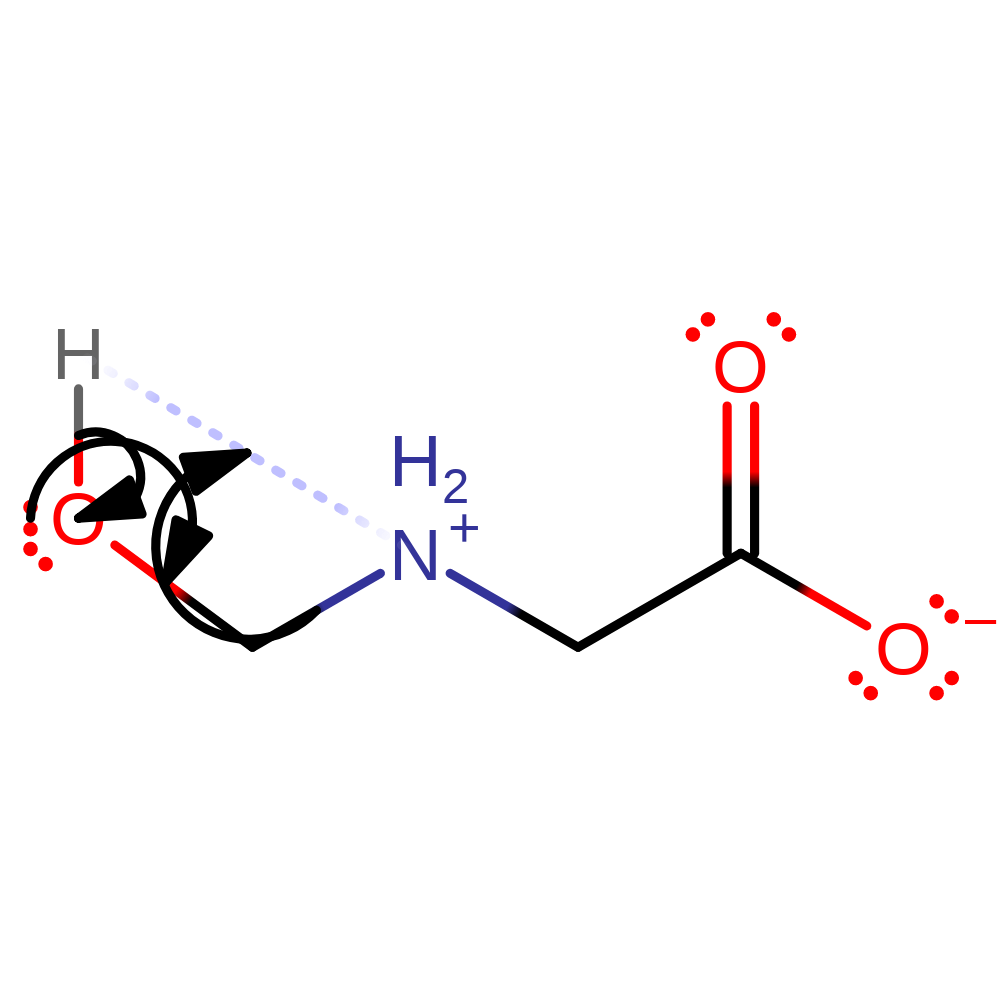
Step 7. The product of the enzyme undergoes spontaneous hydrolysis outside of the active site to produce formaldehyde and glycine.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
reaction occurs outside the enzyme, ingold: intramolecular eliminationIntroduction
The hydride transfer mechanism in which a general base abstracts a proton from the amine group in sarcosine with concomitant transfer of the hydride to the flavin cofactor.
Catalytic Residues Roles
| UniProt | PDB* (2gb0) | ||
| Lys266 | Lys265B | Lys265 is hydrogen bonded to the N(5) position of the flavin ring via a bridging water (wat1) and is also hydrogen bonded to a second nearby water (wat2). The identification of Lys265 as the site of oxygen activation strongly suggests that the solvent molecules might define a pre-organised binding site for the superoxide anion that could accelerate the 1-electron reduction of oxygen by lowering the reorganisation energy associated with transforming the surrounding medium. | proton relay, hydrogen bond donor, proton acceptor, proton donor |
| His270 | His269B | His269 appears to be important in optimising the orientation of bound substrate with respect to electron transfer to flavin. It is also postulated to act as a general acid/base in the polar mechanism. However, mutation of this residue suggests that this second function is less likely. | hydrogen bond donor, proton donor |
| Thr49 | Thr48B | Forms part of a proton relay chain with Lys265 and four water molecules. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, proton relay |
| Cys316 | Cys315B | This residue is covalently attached to the FAD cofactor. This covalent attachment is essential to the enzyme's activity and is thought to function by modulating the redox potential of the FAD and holding the weakly bound FAD in the active site. | covalently attached, activator, alter redox potential |
| Arg50 | Arg49B | The side chain of Arg49 is in van der Waals contact with the si-face of the flavin ring and is essential for covalent flavin attachment. It also plays an important role in sarcosine oxidation by virtue of its electrostatic effect on the active site environment. The positively charged guanidinium side chain in contact with the flavin ring raises the flavin reduction potential and thereby facilitates sarcosine oxidation. An additional role for Arg49 in sarcosine oxidation is suggested by the fact that MSOX is known to bind the unreactive zwitterionic form of its amino acid substrates. Substrate activation is achieved by inducing a large decrease in the pKa of the bound zwitterion. Electrostatic interaction of the reactive substrate anion with the positively charged side chain of Arg49 may contribute to the observed shift in the pKa of the bound amino acid. | modifies pKa, electrostatic stabiliser |
| Lys349, His46 | Lys348B, His45B | Help activate and stabilise the flavin cofactor. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
proton transfer, overall reactant used, cofactor used, intermediate formation, proton relay, hydride transfer, electron transfer, radical formation, inferred reaction step, bimolecular homolytic addition, enzyme-substrate complex formation, aromatic intramolecular elimination, enzyme-substrate complex cleavage, intermediate collapse, intermediate terminated, overall product formed, native state of cofactor regenerated, native state of enzyme regenerated, bimolecular nucleophilic addition, reaction occurs outside the enzyme, intramolecular eliminationReferences
- Trickey P et al. (1999), Structure, 7, 331-345. Monomeric sarcosine oxidase: structure of a covalently flavinylated amine oxidizing enzyme. DOI:10.2210/pdb1l9f/pdb. PMID:10368302.
- Bucci A et al. (2016), J Chem Theory Comput, 12, 2964-2972. Kinetics of O2Entry and Exit in Monomeric Sarcosine Oxidase via Markovian Milestoning Molecular Dynamics. DOI:10.1021/acs.jctc.6b00071. PMID:27168219.
- Pietra F (2015), Chem Biodivers, 12, 1163-1171. On the Quest of Dioxygen by Monomeric Sarcosine Oxidase. A Molecular Dynamics Investigation. DOI:10.1002/cbdv.201400362. PMID:26265568.
- Bucci A et al. (2014), J Chem Theory Comput, 10, 2668-2676. Oxygen Pathways and Allostery in Monomeric Sarcosine Oxidase via Single-Sweep Free-Energy Reconstruction. DOI:10.1021/ct500088z. PMID:25061440.
- Jorns MS et al. (2010), Biochemistry, 49, 3631-3639. Structural Characterization of Mutations at the Oxygen Activation Site in Monomeric Sarcosine Oxidase,. DOI:10.1021/bi100160j. PMID:20353187.
- Hassan-Abdallah A et al. (2008), Biochemistry, 47, 2913-2922. Arginine 49 Is a Bifunctional Residue Important in Catalysis and Biosynthesis of Monomeric Sarcosine Oxidase: A Context-Sensitive Model for the Electrostatic Impact of Arginine to Lysine Mutations†,‡. DOI:10.1021/bi702351v. PMID:18251505.
- Zhao G et al. (2008), Biochemistry, 47, 9124-9135. Identification of the Oxygen Activation Site in Monomeric Sarcosine Oxidase: Role of Lys265 in Catalysis†. DOI:10.1021/bi8008642. PMID:18693755.
- Hassan-Abdallah A et al. (2008), Biochemistry, 47, 1136-1143. Covalent Flavinylation of Monomeric Sarcosine Oxidase: Identification of a Residue Essential for Holoenzyme Biosynthesis†. DOI:10.1021/bi702077q. PMID:18179257.
- Hassan-Abdallah A et al. (2006), Biochemistry, 45, 9454-9462. Role of the Covalent Flavin Linkage in Monomeric Sarcosine Oxidase†. DOI:10.1021/bi0607352. PMID:16878980.
- Zhao G et al. (2006), Biochemistry, 45, 5985-5992. Spectral and Kinetic Characterization of the Michaelis Charge Transfer Complex in Monomeric Sarcosine Oxidase†. DOI:10.1021/bi0600852. PMID:16681370.
- Zhao G et al. (2005), Biochemistry, 44, 16866-16874. Ionization of Zwitterionic Amine Substrates Bound to Monomeric Sarcosine Oxidase†. DOI:10.1021/bi051898d. PMID:16363800.
- Hassan-Abdallah A et al. (2005), Biochemistry, 44, 6452-6462. Biosynthesis of Covalently Bound Flavin: Isolation and in Vitro Flavinylation of the Monomeric Sarcosine Oxidase Apoprotein†. DOI:10.1021/bi047271x. PMID:15850379.
- Khanna P et al. (2003), Biochemistry, 42, 864-869. Tautomeric Rearrangement of a Dihydroflavin Bound to Monomeric Sarcosine Oxidase orN-Methyltryptophan Oxidase†. DOI:10.1021/bi0206098. PMID:12549903.
- Zhao G et al. (2002), Biochemistry, 41, 9747-9750. Monomeric Sarcosine Oxidase: Evidence for an Ionizable Group in the E·S Complex†. DOI:10.1021/bi020285n.
- Zhao G et al. (2002), Biochemistry, 41, 9751-9764. Monomeric Sarcosine Oxidase: Role of Histidine 269 in Catalysis†,‡. DOI:10.1021/bi020286f.
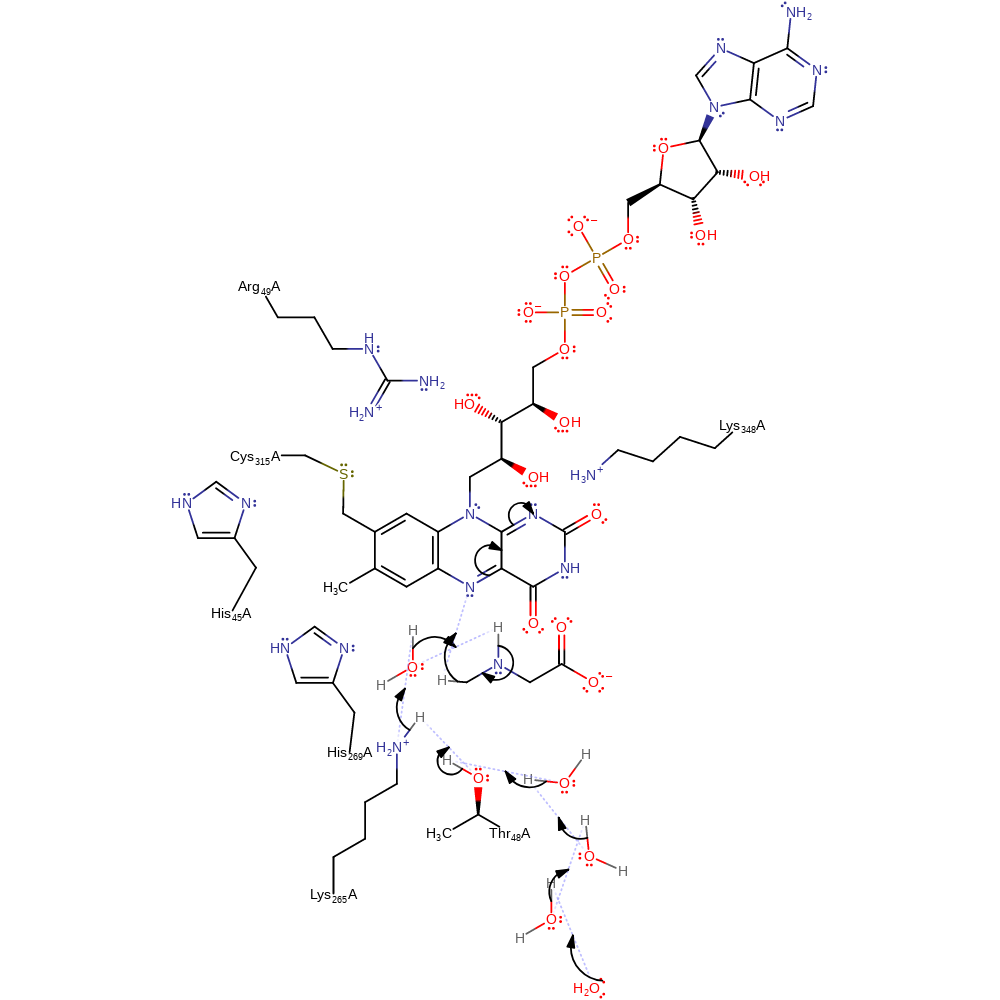
Step 1. A general base (probably the catalytic water via the proton relay chain to bulk solvent) deprotonates the amine group of sarcosine, with concomitant transfer of the hydride to the flavin covator.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His45B | electrostatic stabiliser |
| Arg49B | modifies pKa |
| Cys315B | alter redox potential |
| Lys265B | hydrogen bond donor, proton relay |
| Thr48B | hydrogen bond acceptor, proton relay |
| Cys315B | covalently attached |
| Lys348B | hydrogen bond donor |
| Lys348B | electrostatic stabiliser |
| Thr48B | proton acceptor |
| Lys265B | proton acceptor, proton donor |
| Thr48B | proton donor |
Chemical Components
proton transfer, overall reactant used, cofactor used, intermediate formation, proton relay, hydride transfer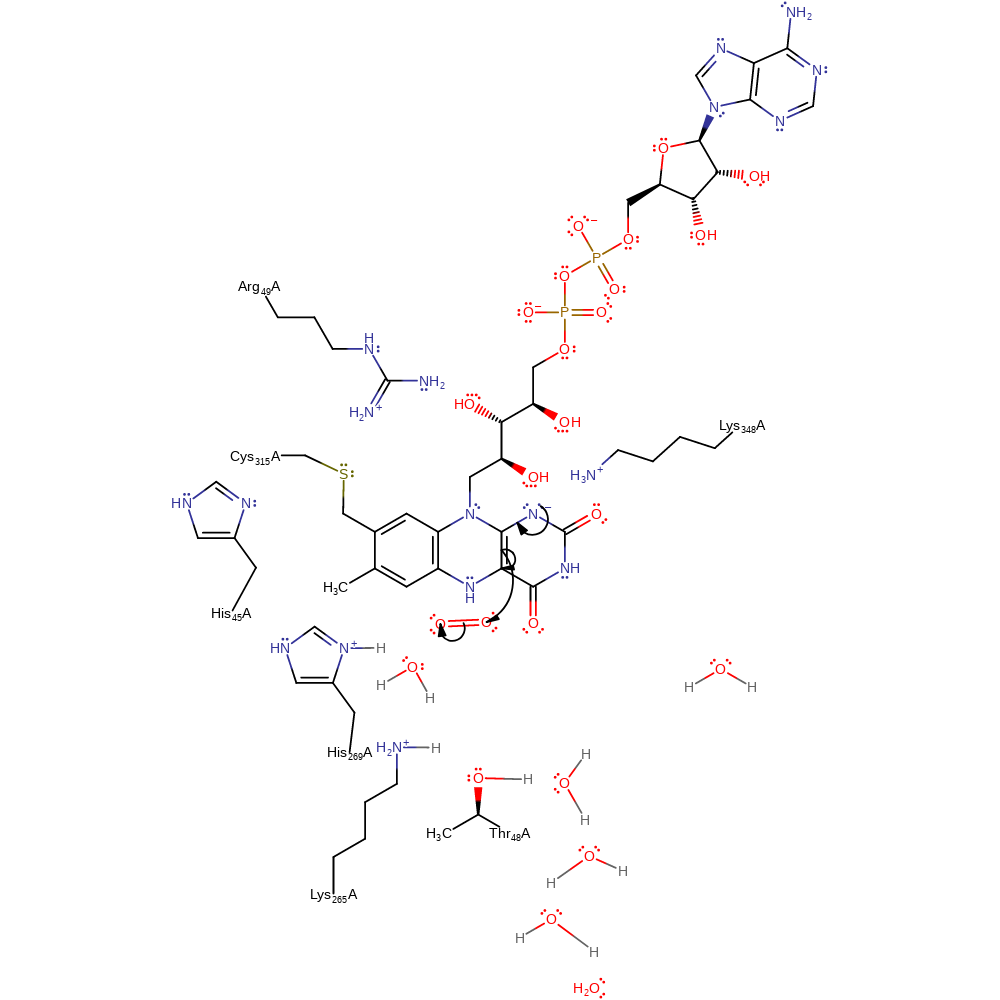
Step 2. The FAD undergoes double bond rearrangement which causes a single electron to be transferred to a dioxygen molecule.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg49B | modifies pKa |
| His45B | electrostatic stabiliser |
| Cys315B | alter redox potential |
| His269B | hydrogen bond donor |
| Lys265B | hydrogen bond donor |
| Thr48B | hydrogen bond acceptor, hydrogen bond donor |
| Cys315B | covalently attached, activator |
| Lys348B | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
electron transfer, radical formation, overall reactant used, intermediate formation, inferred reaction step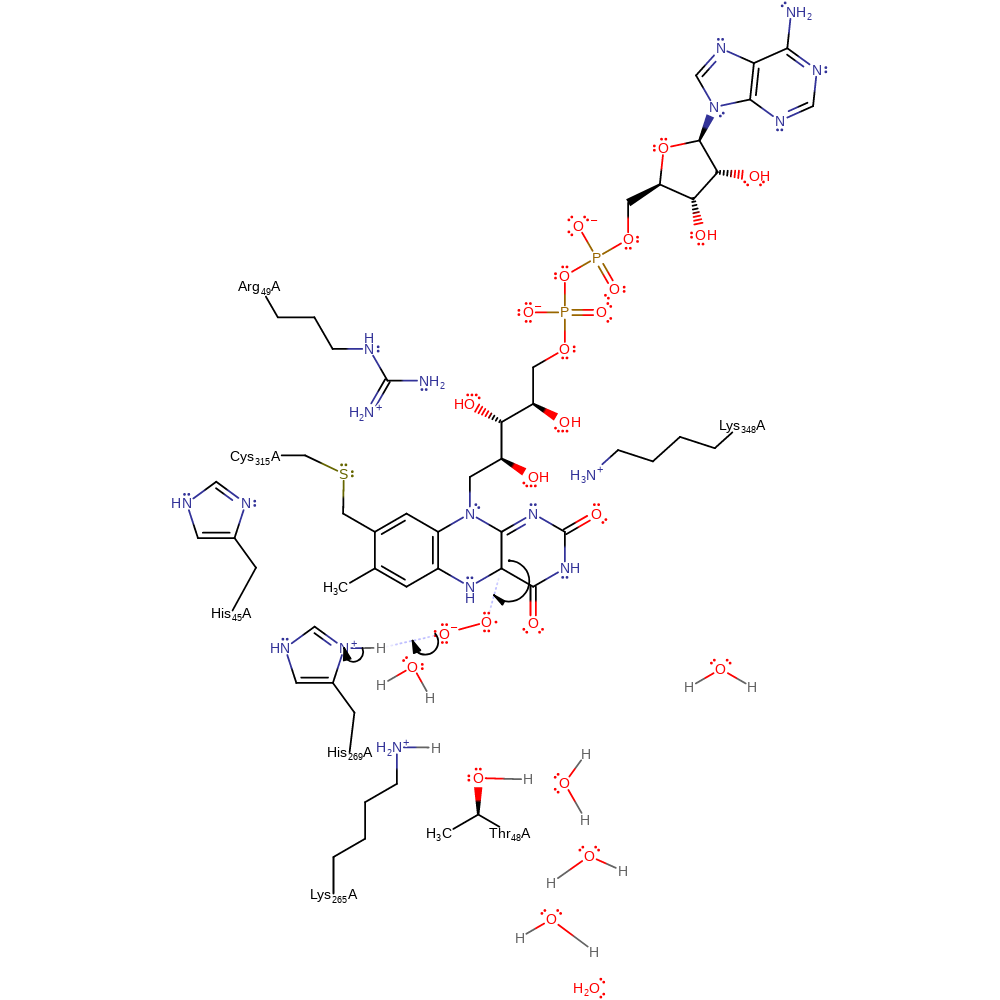
Step 3. The dioxygen molecule undergoes a homolytic reaction in which it colligates to FAD, with concomitant deprotonation of His269.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg49B | electrostatic stabiliser |
| His45B | electrostatic stabiliser |
| Cys315B | alter redox potential |
| His269B | hydrogen bond donor |
| Lys265B | hydrogen bond donor |
| Thr48B | hydrogen bond acceptor, hydrogen bond donor |
| Cys315B | covalently attached, activator |
| Lys348B | hydrogen bond donor |
| His269B | proton donor |
Chemical Components
ingold: bimolecular homolytic addition, proton transfer, enzyme-substrate complex formation, intermediate formation, inferred reaction step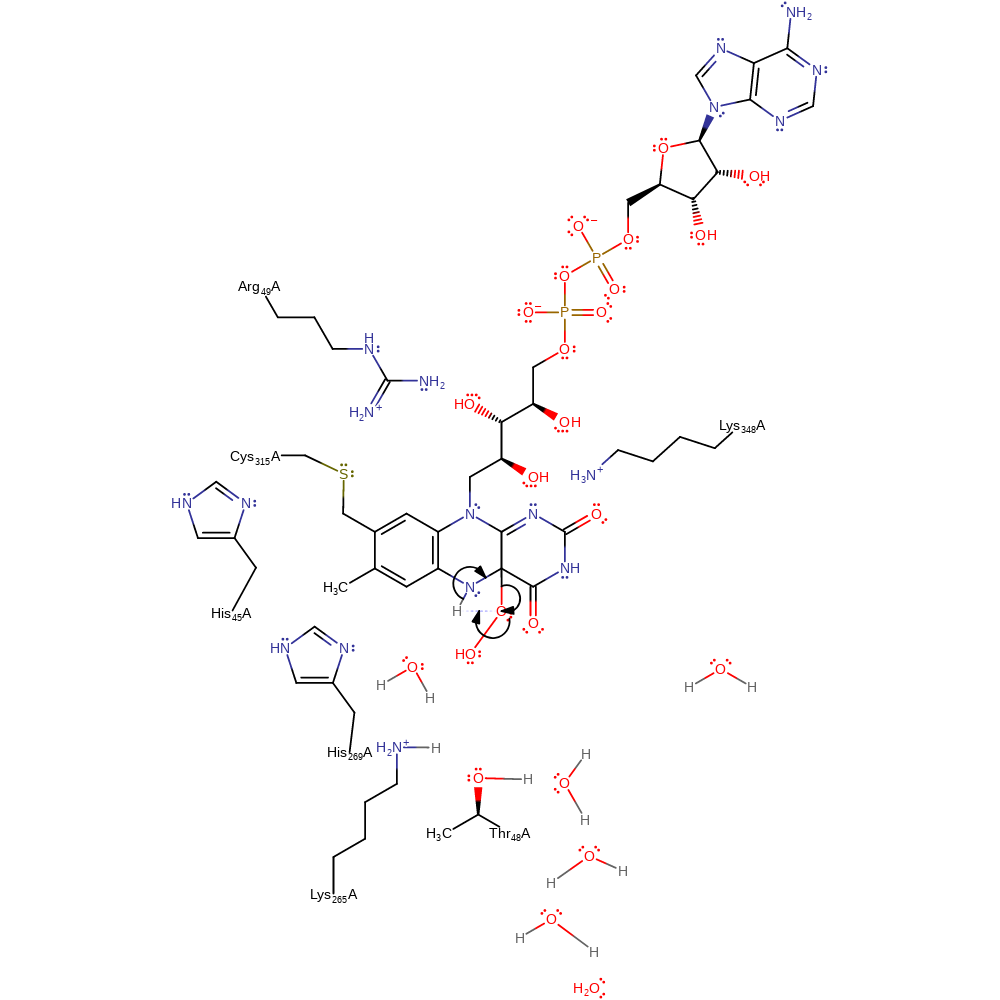
Step 4. The peroxo group deprotonates FAD, which initiates the elimination of hydrogen peroxide.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg49B | electrostatic stabiliser |
| His45B | electrostatic stabiliser |
| Lys265B | hydrogen bond donor |
| Thr48B | hydrogen bond acceptor, hydrogen bond donor |
| Cys315B | covalently attached, activator |
| Lys348B | hydrogen bond donor |
Chemical Components
ingold: aromatic intramolecular elimination, enzyme-substrate complex cleavage, intermediate collapse, intermediate terminated, overall product formed, native state of cofactor regenerated, native state of enzyme regenerated, inferred reaction step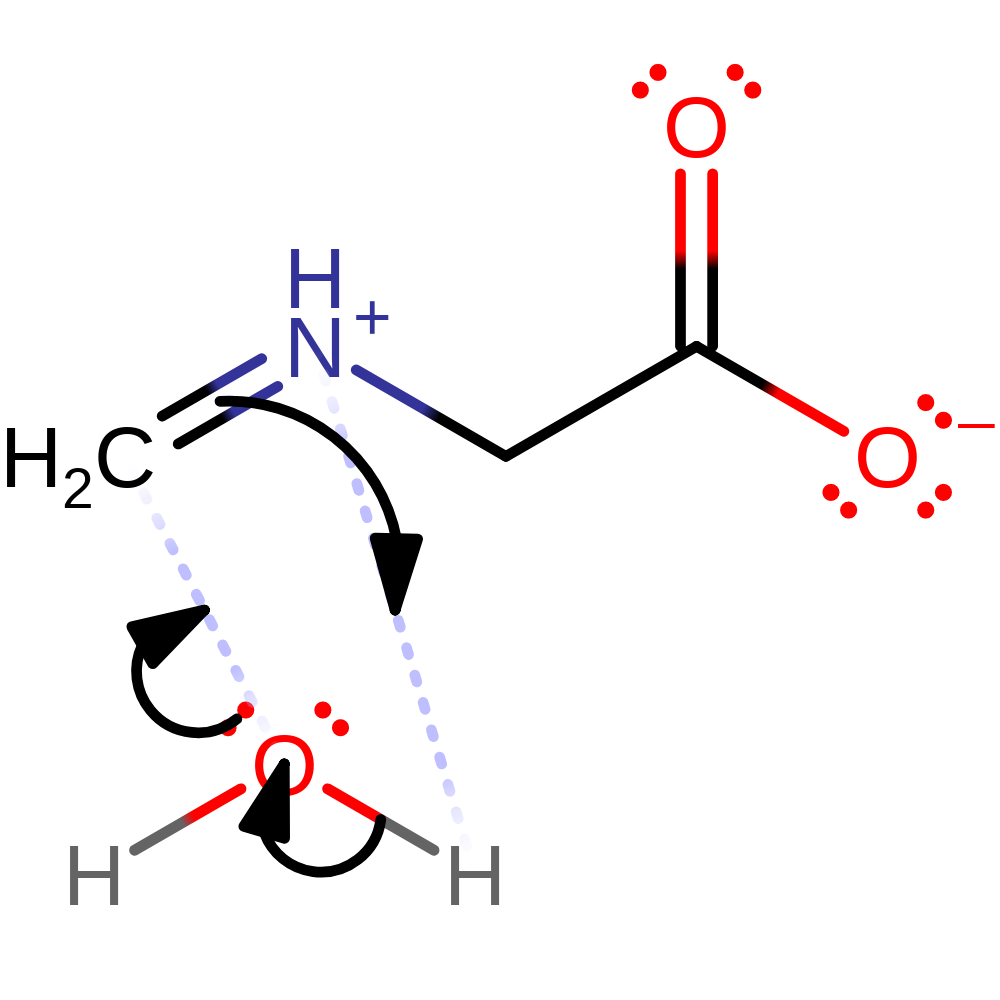
Step 5. The product of the enzyme undergoes spontaneous hydrolysis outside of the active site to produce formaldehyde and glycine.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: bimolecular nucleophilic addition, reaction occurs outside the enzyme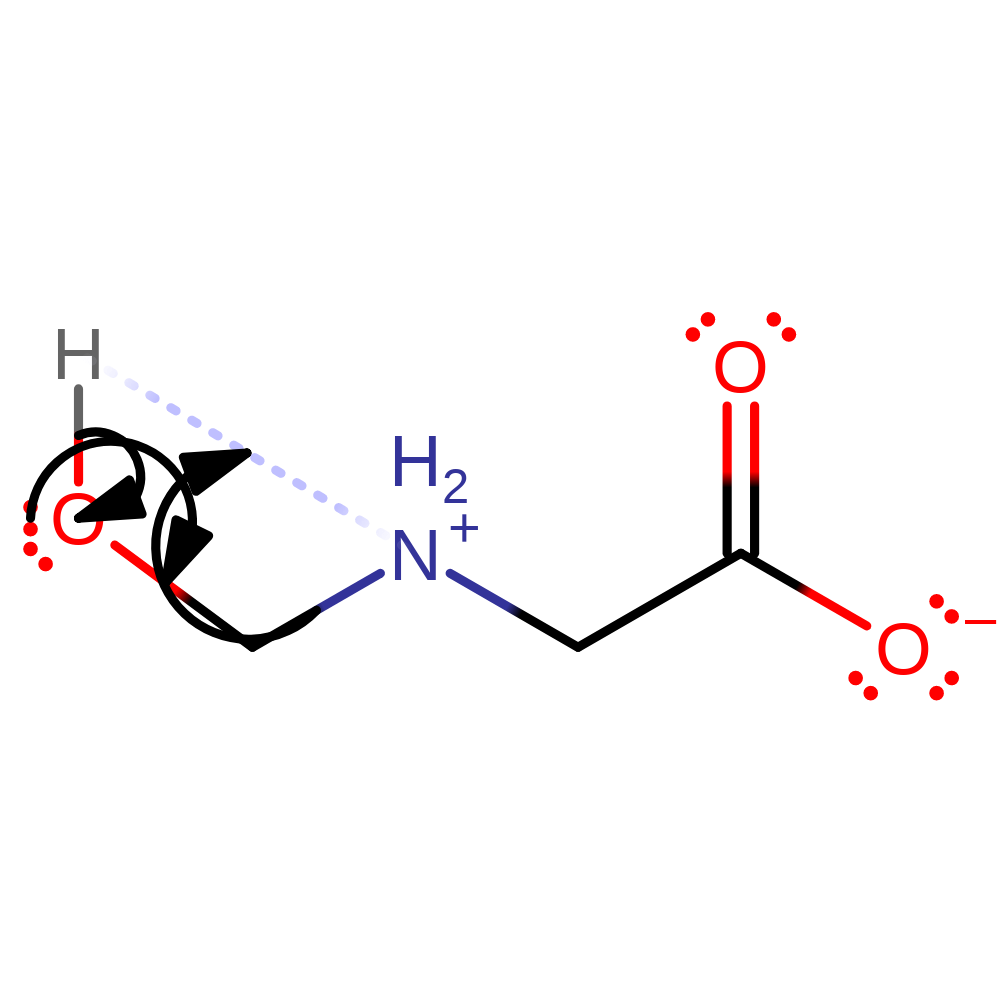
Step 6. The product of the enzyme undergoes spontaneous hydrolysis outside of the active site to produce formaldehyde and glycine.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: intramolecular elimination, reaction occurs outside the enzymeIntroduction
The single electron transfer mechanism in which two single electrons are transferred from the sarcosine to the flavin cofactor.
Catalytic Residues Roles
| UniProt | PDB* (2gb0) | ||
| Lys266 | Lys265B | Lys265 is hydrogen bonded to the N(5) position of the flavin ring via a bridging water (wat1) and is also hydrogen bonded to a second nearby water (wat2). The identification of Lys265 as the site of oxygen activation strongly suggests that the solvent molecules might define a pre-organised binding site for the superoxide anion that could accelerate the 1-electron reduction of oxygen by lowering the reorganisation energy associated with transforming the surrounding medium. | proton relay, hydrogen bond donor, proton acceptor, proton donor |
| His270 | His269B | His269 appears to be important in optimising the orientation of bound substrate with respect to electron transfer to flavin. It is also postulated to act as a general acid/base in the polar mechanism. However, mutation of this residue suggests that this second function is less likely. | hydrogen bond donor, proton acceptor, proton donor |
| Thr49 | Thr48B | Forms part of a proton relay chain with Lys265 and four water molecules. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, proton relay |
| Cys316 | Cys315B | This residue is covalently attached to the FAD cofactor. This covalent attachment is essential to the enzyme's activity and is thought to function by modulating the redox potential of the FAD and holding the weakly bound FAD in the active site. | covalently attached, activator, alter redox potential |
| Arg50 | Arg49B | The side chain of Arg49 is in van der Waals contact with the si-face of the flavin ring and is essential for covalent flavin attachment. It also plays an important role in sarcosine oxidation by virtue of its electrostatic effect on the active site environment. The positively charged guanidinium side chain in contact with the flavin ring raises the flavin reduction potential and thereby facilitates sarcosine oxidation. An additional role for Arg49 in sarcosine oxidation is suggested by the fact that MSOX is known to bind the unreactive zwitterionic form of its amino acid substrates. Substrate activation is achieved by inducing a large decrease in the pKa of the bound zwitterion. Electrostatic interaction of the reactive substrate anion with the positively charged side chain of Arg49 may contribute to the observed shift in the pKa of the bound amino acid. | modifies pKa, electrostatic stabiliser |
| Lys349, His46 | Lys348B, His45B | Help activate and stabilise the flavin cofactor. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
overall reactant used, cofactor used, intermediate formation, electron transfer, proton transfer, radical formation, inferred reaction step, bimolecular homolytic addition, enzyme-substrate complex formation, aromatic intramolecular elimination, enzyme-substrate complex cleavage, intermediate collapse, intermediate terminated, overall product formed, native state of cofactor regenerated, native state of enzyme regenerated, bimolecular nucleophilic addition, reaction occurs outside the enzyme, intramolecular eliminationReferences
- Trickey P et al. (1999), Structure, 7, 331-345. Monomeric sarcosine oxidase: structure of a covalently flavinylated amine oxidizing enzyme. DOI:10.2210/pdb1l9f/pdb. PMID:10368302.
- Bucci A et al. (2016), J Chem Theory Comput, 12, 2964-2972. Kinetics of O2Entry and Exit in Monomeric Sarcosine Oxidase via Markovian Milestoning Molecular Dynamics. DOI:10.1021/acs.jctc.6b00071. PMID:27168219.
- Pietra F (2015), Chem Biodivers, 12, 1163-1171. On the Quest of Dioxygen by Monomeric Sarcosine Oxidase. A Molecular Dynamics Investigation. DOI:10.1002/cbdv.201400362. PMID:26265568.
- Bucci A et al. (2014), J Chem Theory Comput, 10, 2668-2676. Oxygen Pathways and Allostery in Monomeric Sarcosine Oxidase via Single-Sweep Free-Energy Reconstruction. DOI:10.1021/ct500088z. PMID:25061440.
- Jorns MS et al. (2010), Biochemistry, 49, 3631-3639. Structural Characterization of Mutations at the Oxygen Activation Site in Monomeric Sarcosine Oxidase,. DOI:10.1021/bi100160j. PMID:20353187.
- Hassan-Abdallah A et al. (2008), Biochemistry, 47, 2913-2922. Arginine 49 Is a Bifunctional Residue Important in Catalysis and Biosynthesis of Monomeric Sarcosine Oxidase: A Context-Sensitive Model for the Electrostatic Impact of Arginine to Lysine Mutations†,‡. DOI:10.1021/bi702351v. PMID:18251505.
- Zhao G et al. (2008), Biochemistry, 47, 9124-9135. Identification of the Oxygen Activation Site in Monomeric Sarcosine Oxidase: Role of Lys265 in Catalysis†. DOI:10.1021/bi8008642. PMID:18693755.
- Hassan-Abdallah A et al. (2008), Biochemistry, 47, 1136-1143. Covalent Flavinylation of Monomeric Sarcosine Oxidase: Identification of a Residue Essential for Holoenzyme Biosynthesis†. DOI:10.1021/bi702077q. PMID:18179257.
- Hassan-Abdallah A et al. (2006), Biochemistry, 45, 9454-9462. Role of the Covalent Flavin Linkage in Monomeric Sarcosine Oxidase†. DOI:10.1021/bi0607352. PMID:16878980.
- Zhao G et al. (2006), Biochemistry, 45, 5985-5992. Spectral and Kinetic Characterization of the Michaelis Charge Transfer Complex in Monomeric Sarcosine Oxidase†. DOI:10.1021/bi0600852. PMID:16681370.
- Zhao G et al. (2005), Biochemistry, 44, 16866-16874. Ionization of Zwitterionic Amine Substrates Bound to Monomeric Sarcosine Oxidase†. DOI:10.1021/bi051898d. PMID:16363800.
- Hassan-Abdallah A et al. (2005), Biochemistry, 44, 6452-6462. Biosynthesis of Covalently Bound Flavin: Isolation and in Vitro Flavinylation of the Monomeric Sarcosine Oxidase Apoprotein†. DOI:10.1021/bi047271x. PMID:15850379.
- Khanna P et al. (2003), Biochemistry, 42, 864-869. Tautomeric Rearrangement of a Dihydroflavin Bound to Monomeric Sarcosine Oxidase orN-Methyltryptophan Oxidase†. DOI:10.1021/bi0206098. PMID:12549903.
- Zhao G et al. (2002), Biochemistry, 41, 9747-9750. Monomeric Sarcosine Oxidase: Evidence for an Ionizable Group in the E·S Complex†. DOI:10.1021/bi020285n.
- Zhao G et al. (2002), Biochemistry, 41, 9751-9764. Monomeric Sarcosine Oxidase: Role of Histidine 269 in Catalysis†,‡. DOI:10.1021/bi020286f.
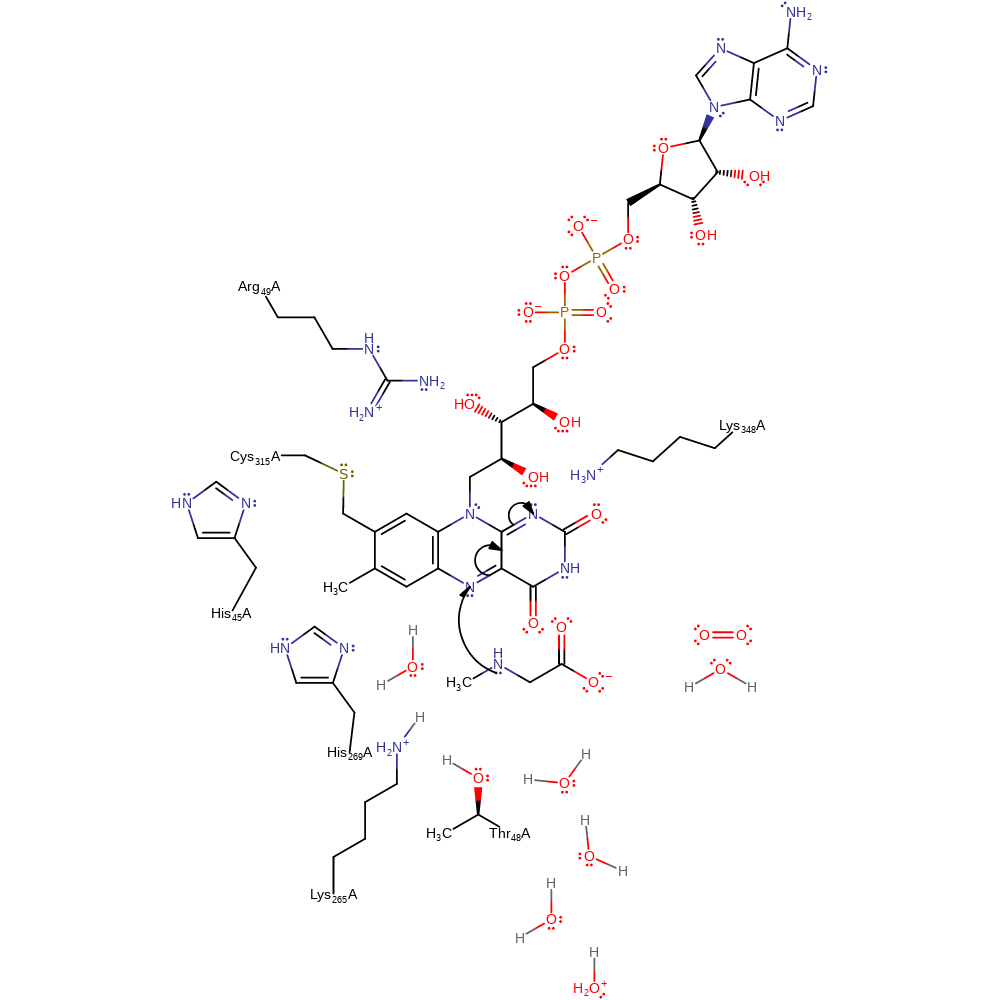
Step 1. A single electron is transferred from the sarcosine to the flavin cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His45B | electrostatic stabiliser |
| Arg49B | modifies pKa |
| Cys315B | alter redox potential |
| Lys348B | hydrogen bond donor |
| Cys315B | covalently attached |
| Lys348B | electrostatic stabiliser |
Chemical Components
overall reactant used, cofactor used, intermediate formation, electron transfer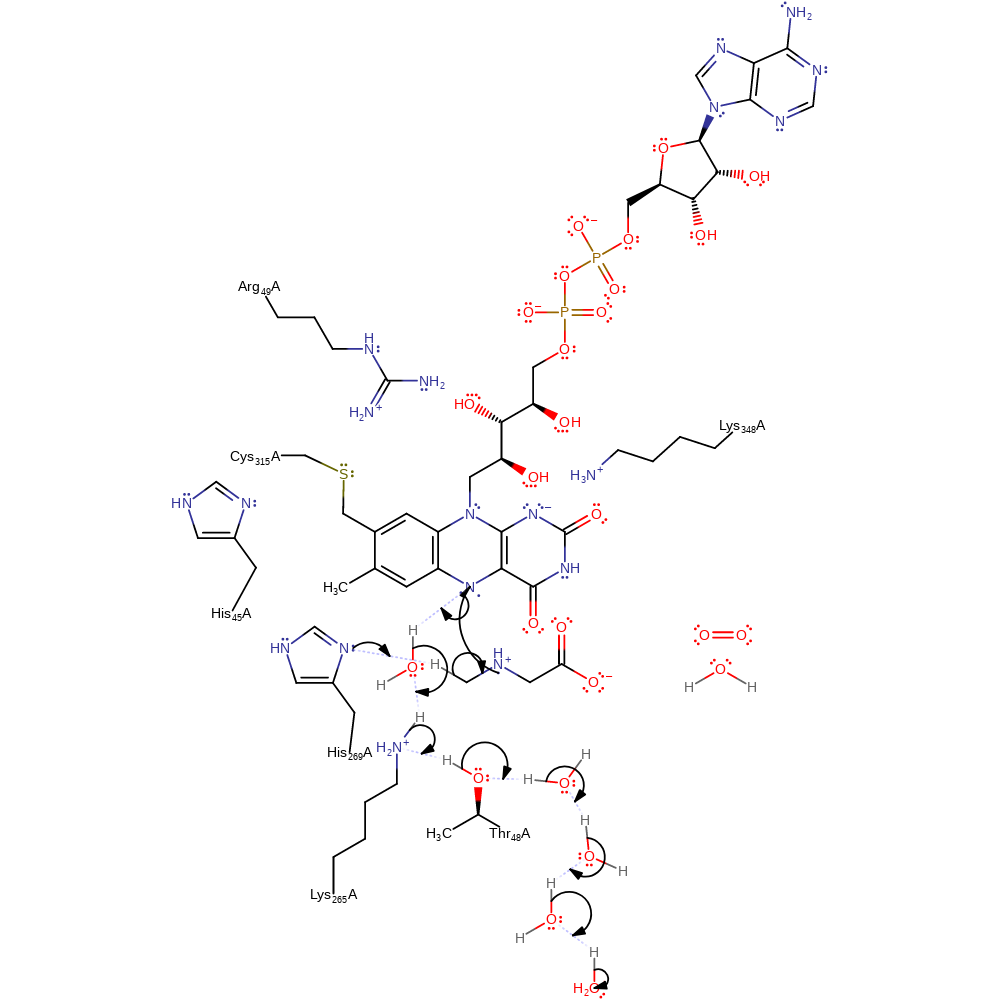
Step 2. His269 deprotonates the methyl group, initiating a second single electron transfer to the flavin cofactor and abstraction of a proton from bulk solvent via the proton relay.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg49B | modifies pKa |
| Lys348B | electrostatic stabiliser |
| Thr48B | proton acceptor, proton relay |
| Lys265B | proton relay |
| His269B | proton acceptor |
| Lys265B | proton donor, proton acceptor |
| Thr48B | proton donor |
Chemical Components
proton transfer, electron transfer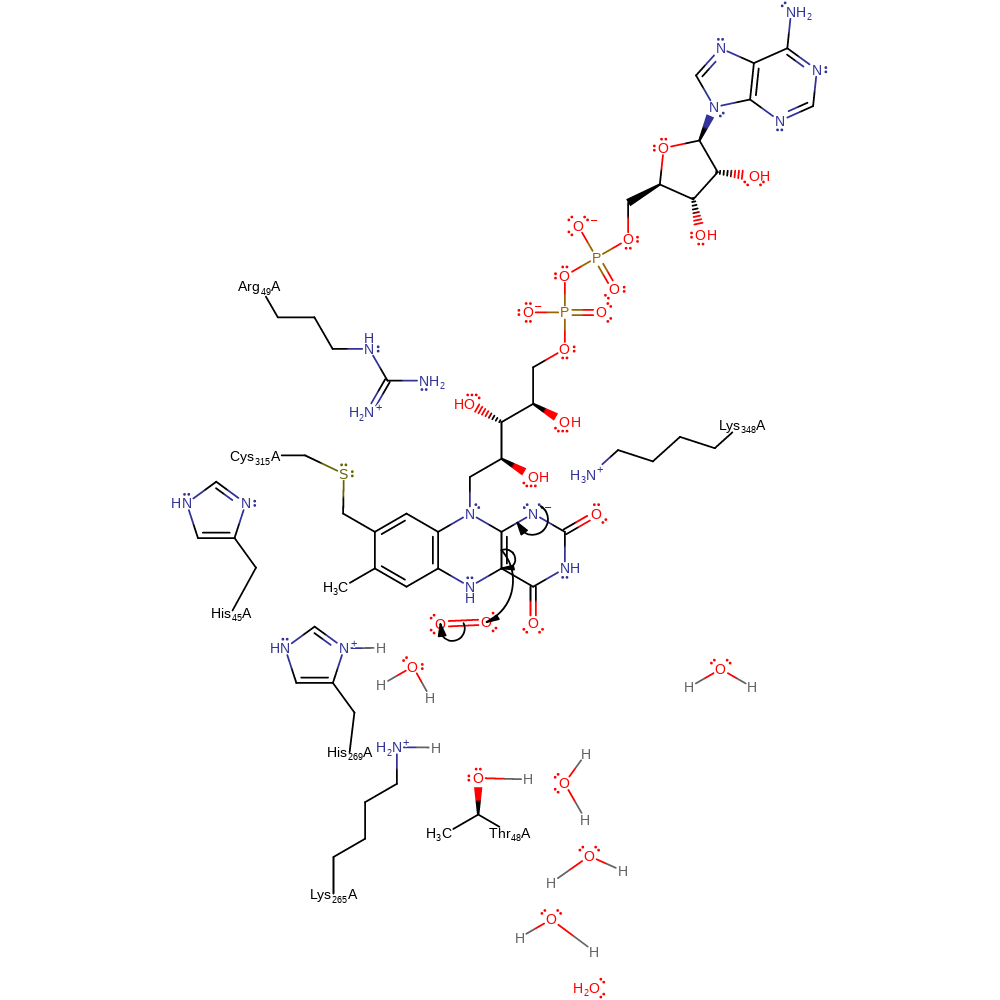
Step 3. The FAD undergoes double bond rearrangement which causes a single electron to be transferred to a dioxygen molecule.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg49B | modifies pKa |
| His45B | electrostatic stabiliser |
| Cys315B | alter redox potential |
| His269B | hydrogen bond donor |
| Lys265B | hydrogen bond donor |
| Thr48B | hydrogen bond acceptor, hydrogen bond donor |
| Cys315B | covalently attached, activator |
| Lys348B | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
electron transfer, radical formation, overall reactant used, intermediate formation, inferred reaction step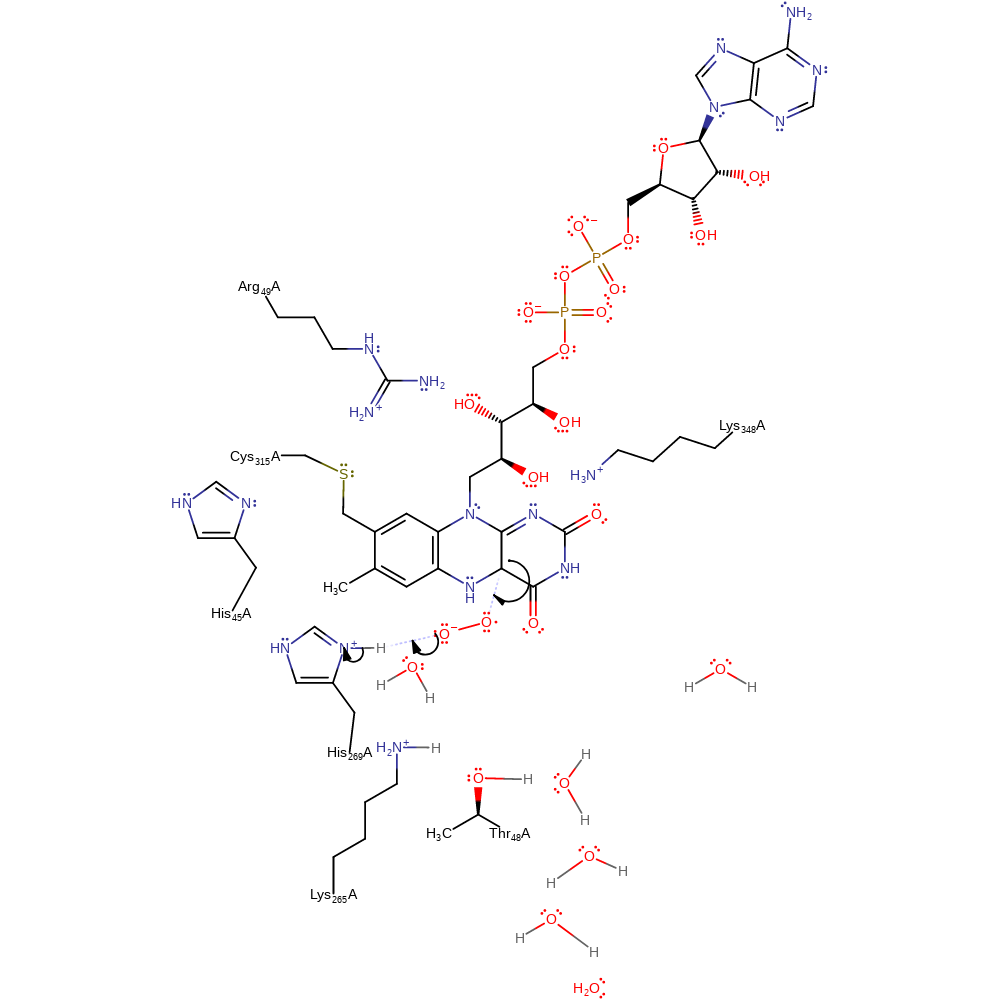
Step 4. The dioxygen molecule undergoes a homolytic reaction in which it colligates to FAD, with concomitant deprotonation of His269.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg49B | electrostatic stabiliser |
| His45B | electrostatic stabiliser |
| Cys315B | alter redox potential |
| His269B | hydrogen bond donor |
| Lys265B | hydrogen bond donor |
| Thr48B | hydrogen bond acceptor, hydrogen bond donor |
| Cys315B | covalently attached, activator |
| Lys348B | hydrogen bond donor |
| His269B | proton donor |
Chemical Components
ingold: bimolecular homolytic addition, proton transfer, enzyme-substrate complex formation, intermediate formation, inferred reaction step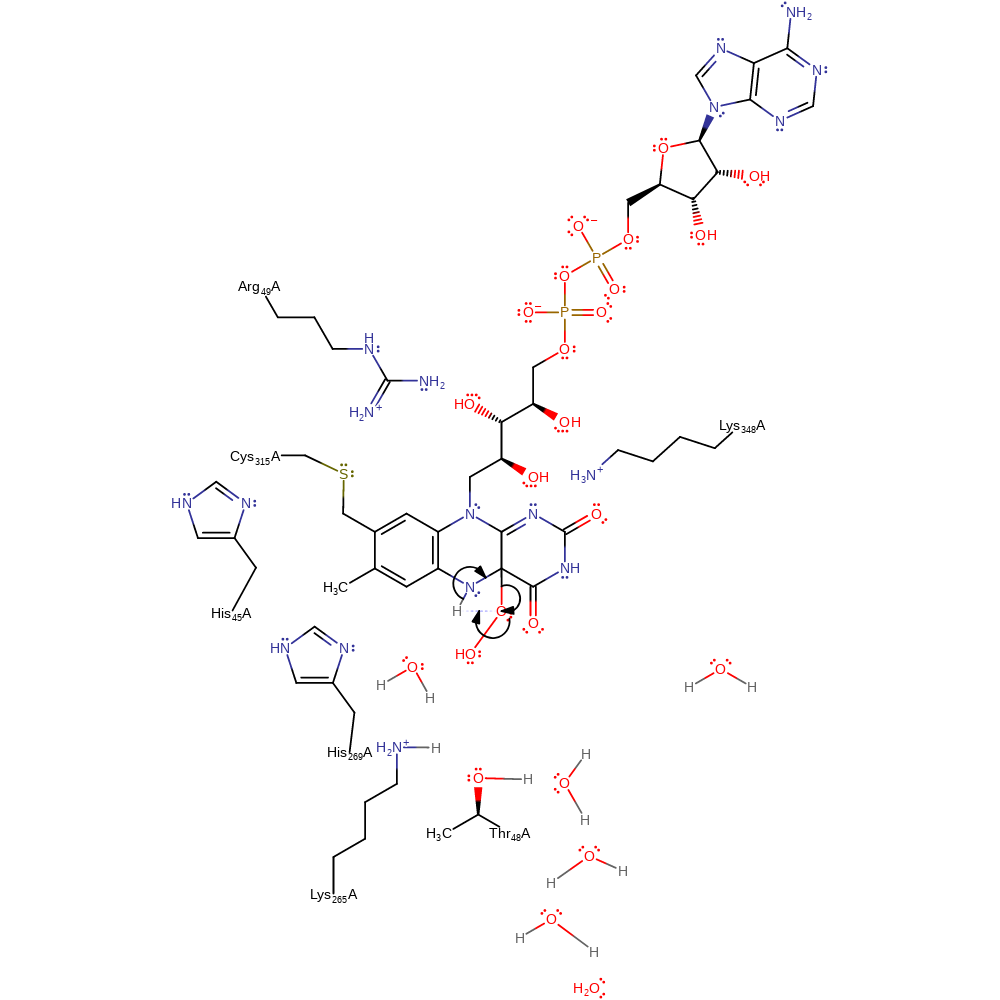
Step 5. The peroxo group deprotonates FAD, which initiates the elimination of hydrogen peroxide.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg49B | electrostatic stabiliser |
| His45B | electrostatic stabiliser |
| Lys265B | hydrogen bond donor |
| Thr48B | hydrogen bond acceptor, hydrogen bond donor |
| Cys315B | covalently attached, activator |
| Lys348B | hydrogen bond donor |
Chemical Components
ingold: aromatic intramolecular elimination, enzyme-substrate complex cleavage, intermediate collapse, intermediate terminated, overall product formed, native state of cofactor regenerated, native state of enzyme regenerated, inferred reaction step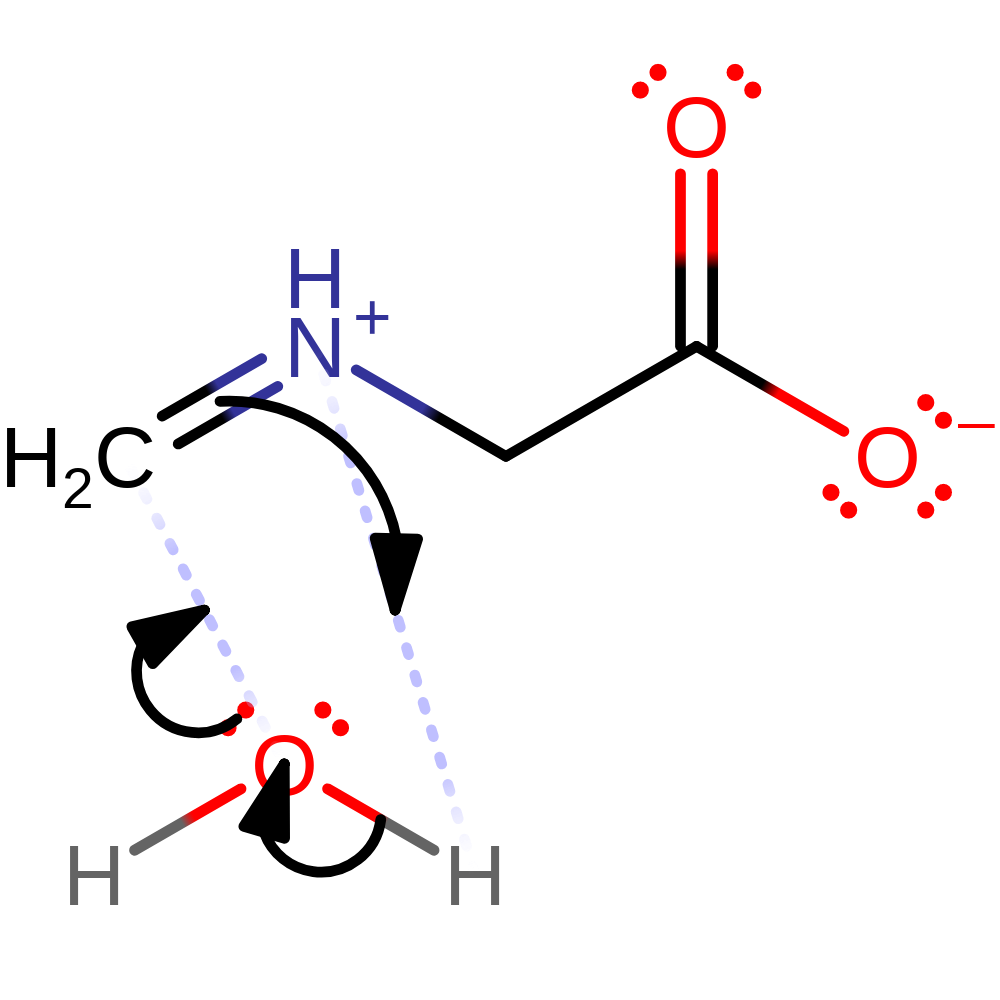
Step 6. The product of the enzyme undergoes spontaneous hydrolysis outside of the active site to produce formaldehyde and glycine.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: bimolecular nucleophilic addition, reaction occurs outside the enzyme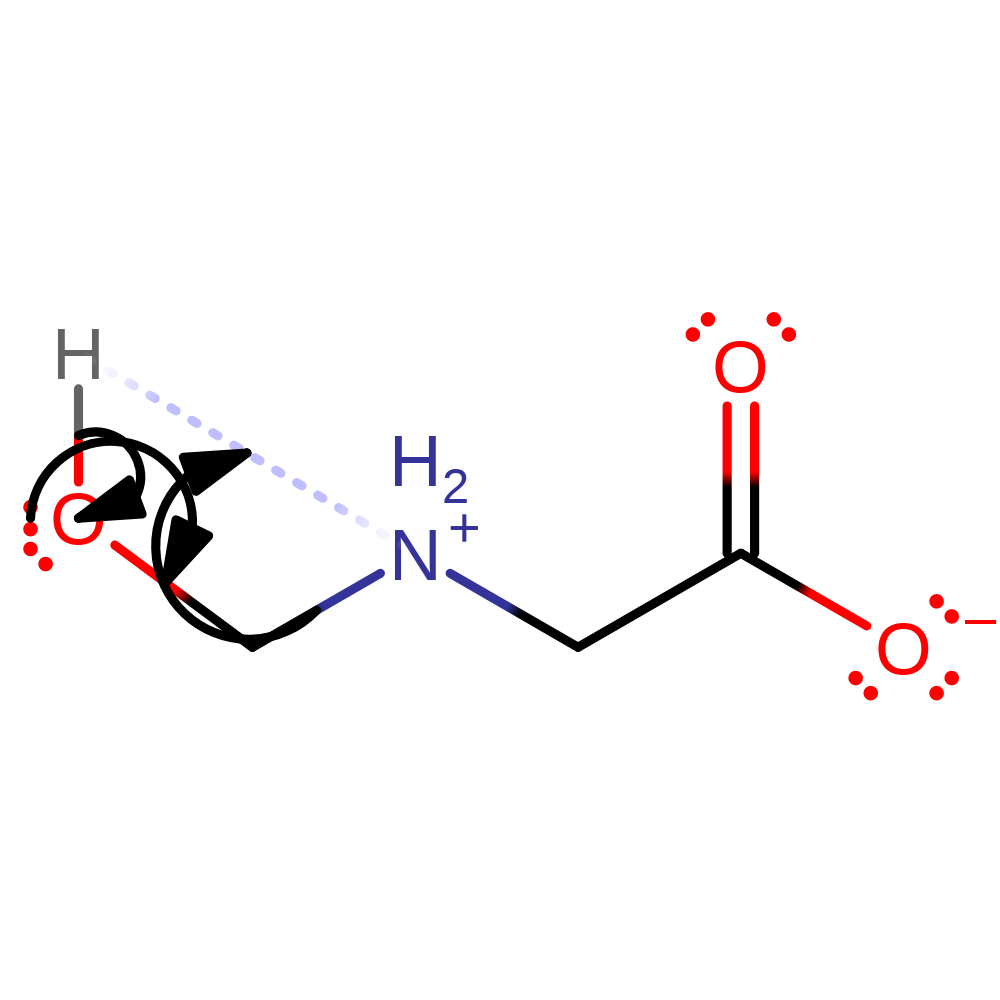
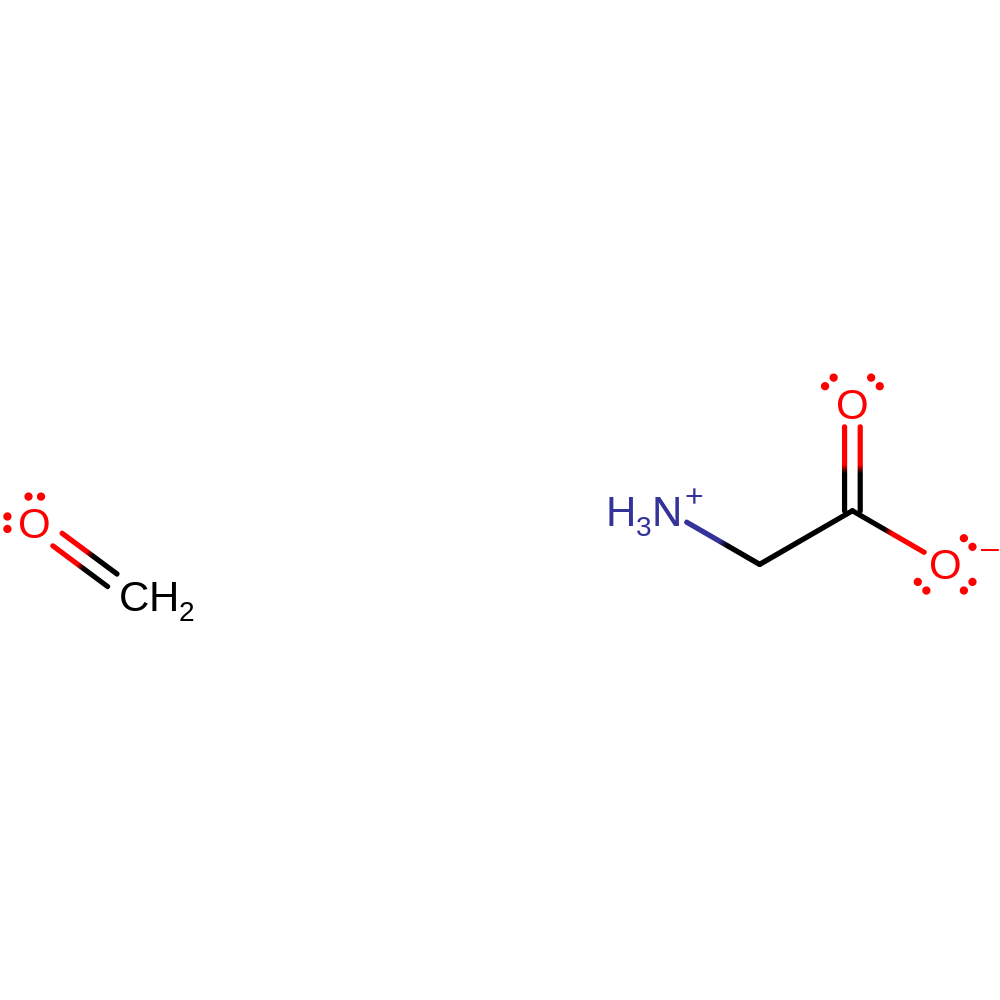
Step 7. The product of the enzyme undergoes spontaneous hydrolysis outside of the active site to produce formaldehyde and glycine.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|






 Download:
Download: 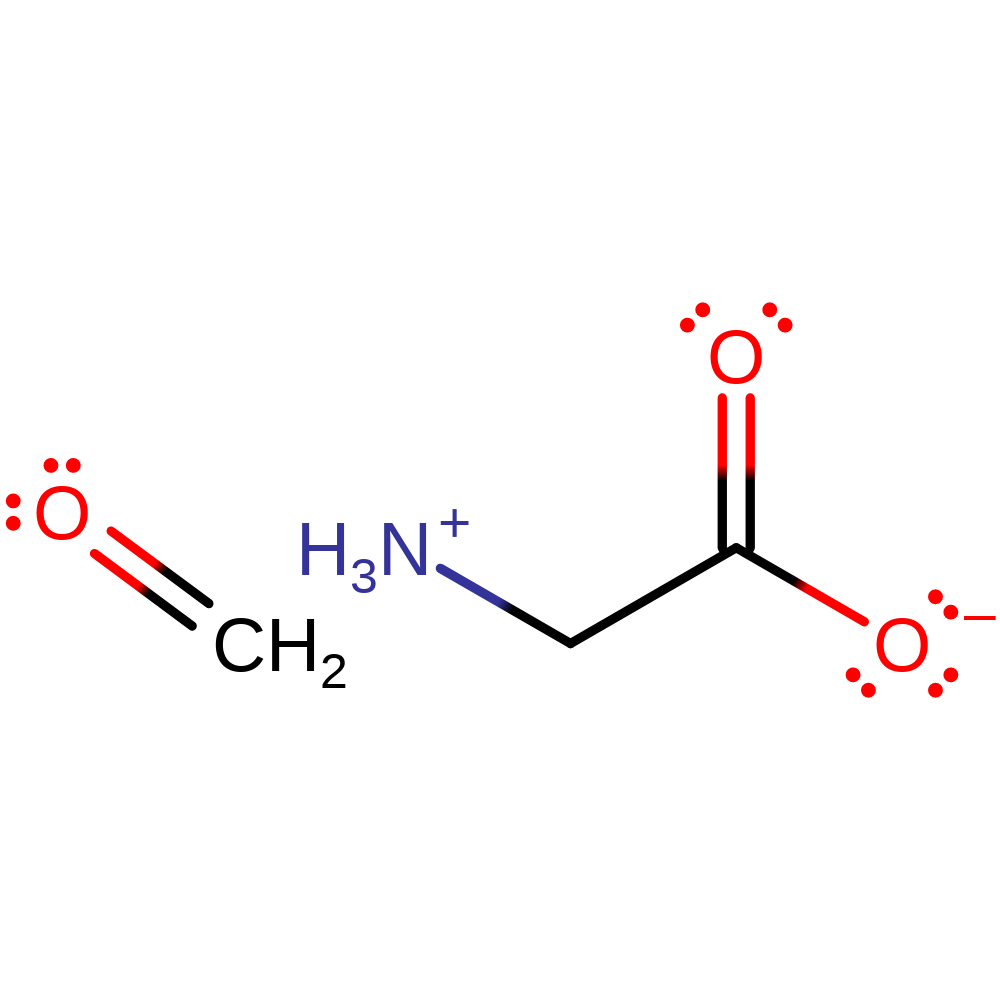 Download:
Download: 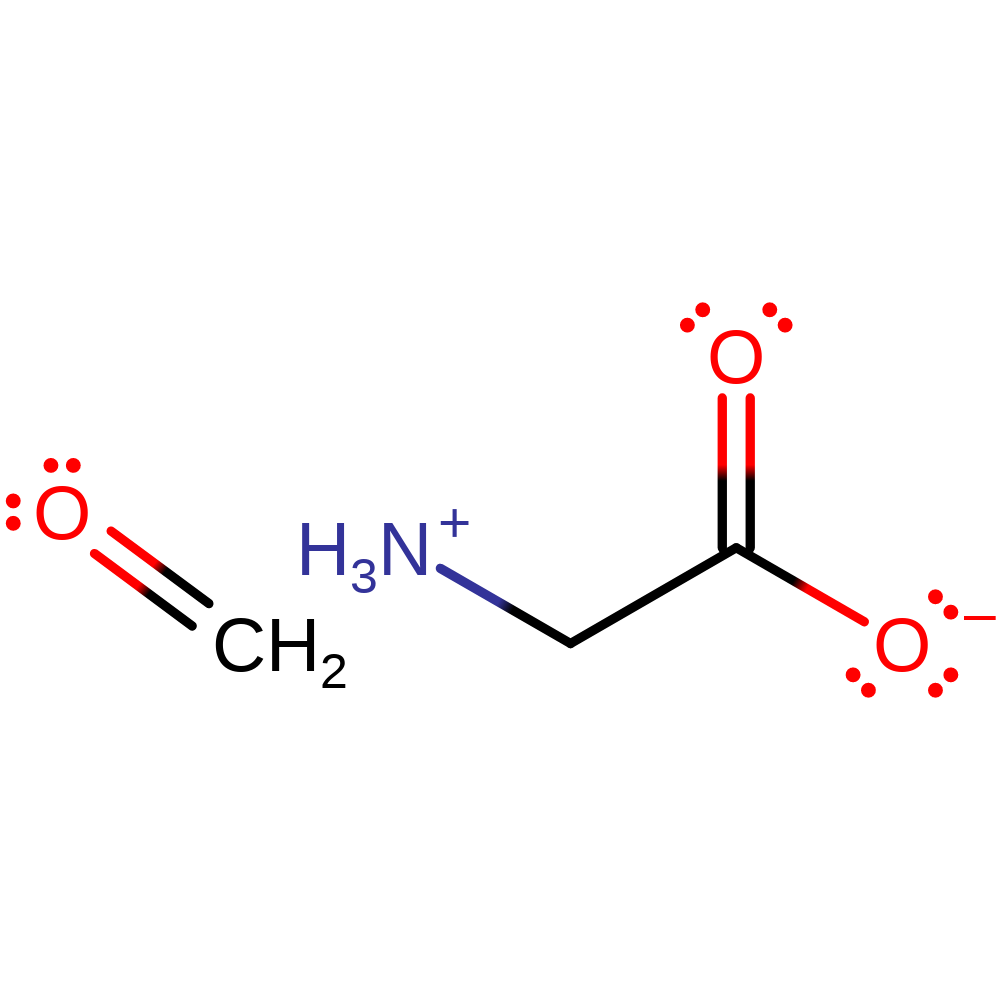 Download:
Download: