D-amino-acid oxidase
D amino acid oxidase (DAAO) acts to oxidise D amino acids to their imino counterparts, using FAD as a cofactor. The imino acid then undergoes hydrolysis outside of the enzyme's active site. This reaction has been found to play an important role in the degradation of certain neurotransmitters such as D-serine in mammals, but the enzyme is also found in other eukaryotes and its function has not been fully characterised. It shows mechanistic correlation with other enzymes able to oxidise amino groups.
Yeast and mammalian DAAOs share features such as the basic catalytic mechanism. However, they differ in important aspects such as catalytic efficiency, substrate specificity, aggregation state, stability, kinetic mechanism, and mode and effectiveness of FAD binding. Thus, DAAO from the yeast Rhodotorula gracilis (RgDAAO) has a kcat value ≈20,000 minute−1 compared to 600 minute−1 for pig kidney DAAO (pkDAAO) with D-alanine as substrate.
Reference Protein and Structure
- Sequence
-
P80324
 (1.4.3.3)
(1.4.3.3)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Rhodotorula toruloides (Yeast)

- PDB
-
1c0p
- D-AMINO ACIC OXIDASE IN COMPLEX WITH D-ALANINE AND A PARTIALLY OCCUPIED BIATOMIC SPECIES
(1.2 Å)



- Catalytic CATH Domains
-
3.40.50.720
 (see all for 1c0p)
(see all for 1c0p)
- Cofactors
- Fadh2(2-) (1)
Enzyme Reaction (EC:1.4.3.3)
Enzyme Mechanism
Introduction
The mechanism of the reaction is through direct hydride transfer from the alpha carbon of the amino acid to the N5 of the FAD, with concomitant deprotonation of the alpha amino group by Ser 335's side chain allowing orbital overlap between the nitrogen lone pair and the carbon's sigma* orbital to occur, resulting in the imine product. The transition state for this process is planar and stabilised by hydrogen bonding between the carbonyl of Ser 335 and the amino group. This mechanism is supported by high resolution crystal structures, kinetic isotope effects and free energy correlation calculations [PMID:11070076].
Catalytic Residues Roles
| UniProt | PDB* (1c0p) | ||
| Ser335 (main-C), Gln339 (main-C), Asn54 (main-N), Asn54 (main-C) | Ser1335(337)A (main-C), Gln1339(341)A (main-C), Asn1054(56)A (main-N), Asn1054(56)A (main-C) | Acts to lower the pKa of the amino acid substrate. | modifies pKa, hydrogen bond acceptor, hydrogen bond donor |
| Ser335 | Ser1335(337)A | Acts as a general acid/base. | proton relay, proton acceptor, proton donor |
Chemical Components
bimolecular elimination, hydride transfer, aromatic bimolecular nucleophilic addition, overall reactant used, cofactor used, intermediate formation, overall product formed, proton relay, rate-determining step, radical formation, electron transfer, radical termination, bimolecular homolytic addition, proton transfer, aromatic intramolecular elimination, native state of cofactor regenerated, intermediate terminated, native state of enzyme regenerated, reaction occurs outside the enzyme, bimolecular nucleophilic addition, deamination, intramolecular eliminationReferences
- Umhau S et al. (2000), Proc Natl Acad Sci U S A, 97, 12463-12468. The x-ray structure of D-amino acid oxidase at very high resolution identifies the chemical mechanism of flavin-dependent substrate dehydrogenation. DOI:10.1073/pnas.97.23.12463. PMID:11070076.
- Ghisla S et al. (2011), J Biol Chem, 286, 40987-40998. Revisitation of the Cl-Elimination Reaction of D-Amino Acid Oxidase: NEW INTERPRETATION OF THE REACTION THAT SPARKED FLAVOPROTEIN DEHYDROGENATION MECHANISMS. DOI:10.1074/jbc.m111.266536. PMID:21949129.
- Rosini E et al. (2011), FEBS J, 278, 482-492. On the reaction of d-amino acid oxidase with dioxygen: O2 diffusion pathways and enhancement of reactivity. DOI:10.1111/j.1742-4658.2010.07969.x. PMID:21182588.
- Katane M et al. (2008), Amino Acids, 35, 75-82. Hyperactive mutants of mouse d-aspartate oxidase: mutagenesis of the active site residue serine 308. DOI:10.1007/s00726-007-0627-8. PMID:18235994.
- Boselli A et al. (2007), Biochimie, 89, 360-368. Investigating the role of active site residues of Rhodotorula gracilis d-amino acid oxidase on its substrate specificity. DOI:10.1016/j.biochi.2006.10.017. PMID:17145127.
- Caldinelli L et al. (2006), FEBS J, 273, 504-512. Tryptophan 243 affects interprotein contacts, cofactor binding and stability in D-amino acid oxidase from Rhodotorula gracilis. DOI:10.1111/j.1742-4658.2005.05083.x. PMID:16420474.
- Tishkov VI et al. (2005), Biochemistry (Mosc), 70, 40-54. D-amino acid oxidase: structure, catalytic mechanism, and practical application. DOI:10.1007/s10541-005-0050-2.
- Boselli A et al. (2004), Biochim Biophys Acta, 1702, 19-32. On the mechanism of Rhodotorula gracilis d-amino acid oxidase: role of the active site serine 335. DOI:10.1016/j.bbapap.2004.07.005. PMID:15450847.
- Pollegioni L et al. (2004), Biotechnol Prog, 20, 467-473. Catalytic Properties of d-Amino Acid Oxidase in Cephalosporin C Bioconversion: A Comparison between Proteins from Different Sources. DOI:10.1021/bp034206q. PMID:15058991.
- Pollegioni L et al. (2002), J Mol Biol, 324, 535-546. Yeast d-Amino Acid Oxidase: Structural Basis of its Catalytic Properties. DOI:10.1016/s0022-2836(02)01062-8.
- Pilone MS (2000), Cell Mol Life Sci, 57, 1732-1747. D-Amino acid oxidase: new findings. DOI:10.1007/pl00000655. PMID:11130179.
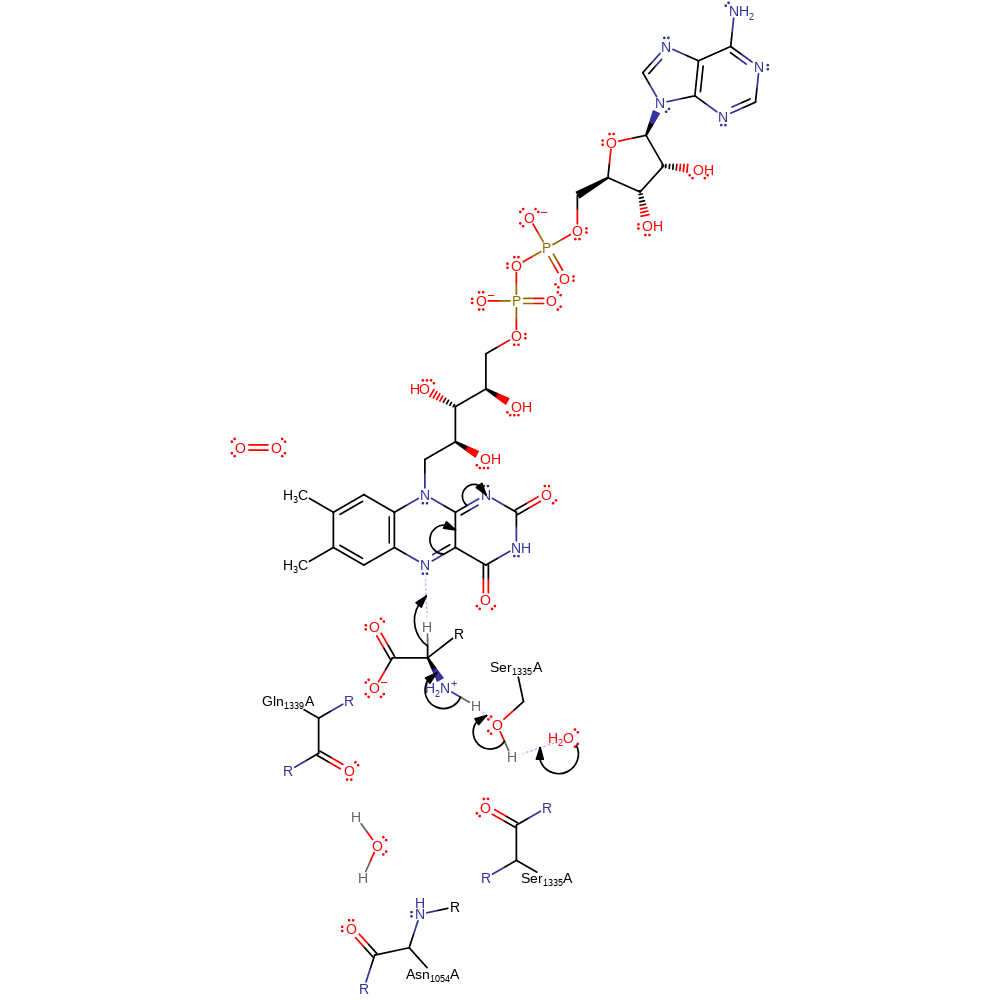
Step 1. Water deprotonates Ser1335, which deprotonates the amine of the D-amino acid, eliminating a hydride ion, which adds to FAD.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser1335(337)A (main-C) | hydrogen bond acceptor, hydrogen bond donor |
| Asn1054(56)A (main-N) | hydrogen bond donor, hydrogen bond acceptor |
| Gln1339(341)A (main-C) | hydrogen bond acceptor |
| Asn1054(56)A (main-N) | modifies pKa |
| Asn1054(56)A (main-C) | modifies pKa |
| Ser1335(337)A (main-C) | modifies pKa |
| Gln1339(341)A (main-C) | modifies pKa |
| Ser1335(337)A | proton donor, proton relay, proton acceptor |
Chemical Components
ingold: bimolecular elimination, hydride transfer, ingold: aromatic bimolecular nucleophilic addition, overall reactant used, cofactor used, intermediate formation, overall product formed, proton relay, rate-determining step
Step 2. The FAD undergoes double bond rearrangement which causes a single electron to be transferred to a dioxygen molecule.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
radical formation, electron transfer, overall reactant used, intermediate formation
Step 3. The dioxygen molecule undergoes a homolytic reaction in which it colligates to FAD, with concomitant deprotonation of water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn1054(56)A (main-N) | hydrogen bond donor, hydrogen bond acceptor |
| Gln1339(341)A (main-C) | hydrogen bond acceptor |
Chemical Components
radical termination, ingold: bimolecular homolytic addition, proton transfer, intermediate formation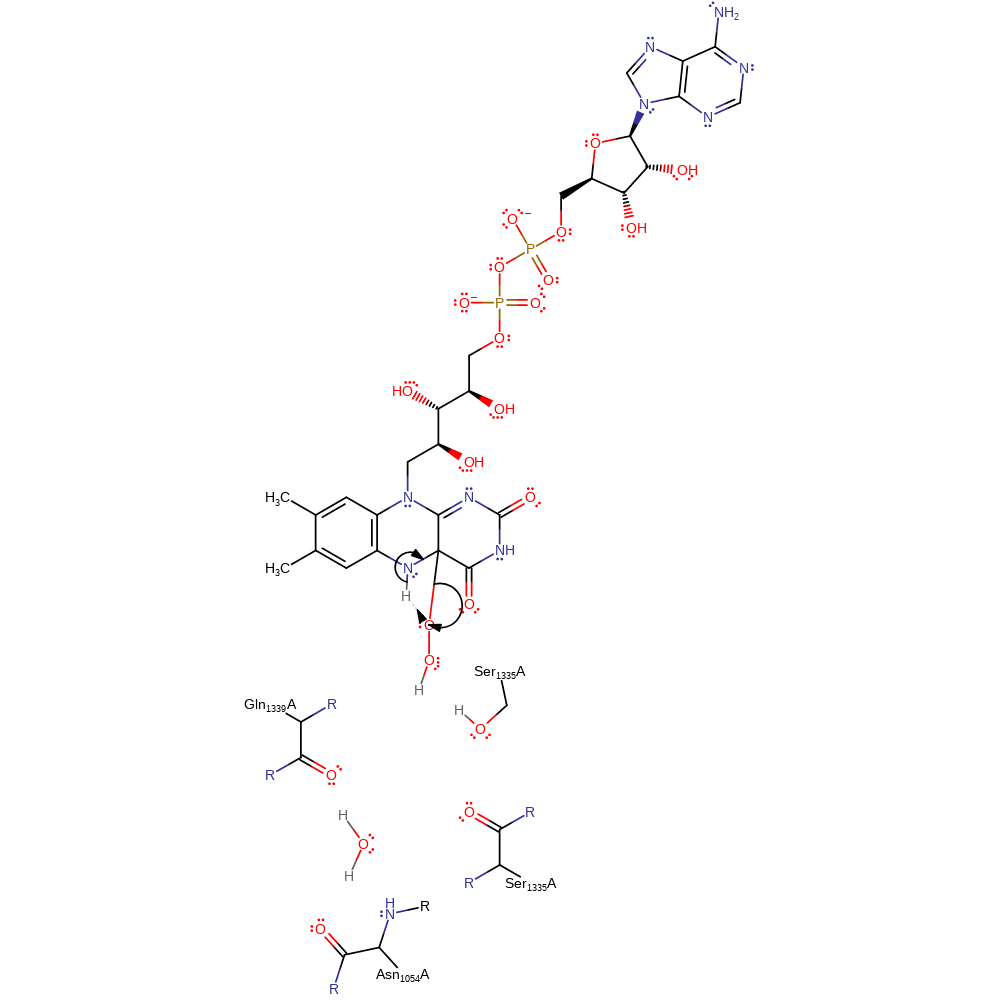
Step 4. The peroxo group deprotonates FAD, which initiates the elimination of hydrogen peroxide.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn1054(56)A (main-N) | hydrogen bond donor, hydrogen bond acceptor |
| Gln1339(341)A (main-C) | hydrogen bond acceptor |
Chemical Components
ingold: aromatic intramolecular elimination, native state of cofactor regenerated, intermediate terminated, overall product formed, native state of enzyme regenerated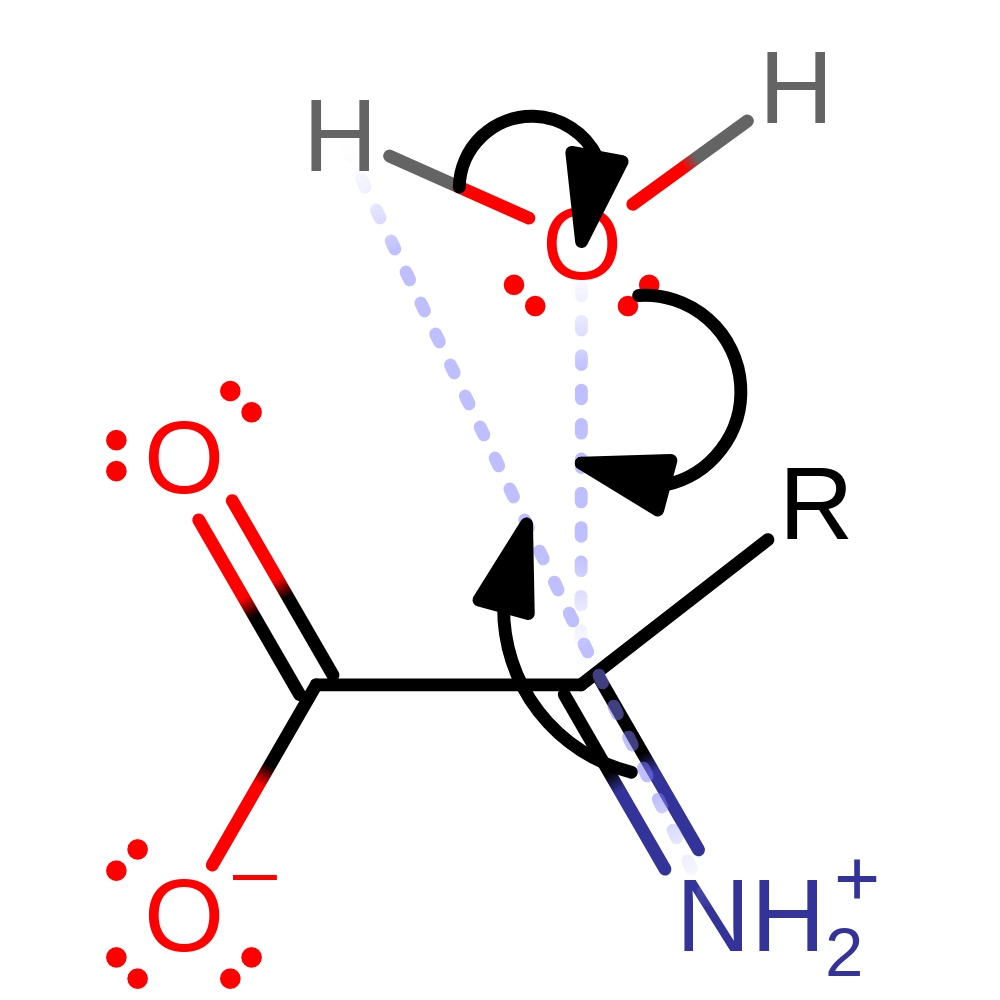
Step 5. The product of the enzyme undergoes spontaneous hydrolysis outside of the active site to produce ammonium and the 2-oxo acid.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
reaction occurs outside the enzyme, proton transfer, ingold: bimolecular nucleophilic addition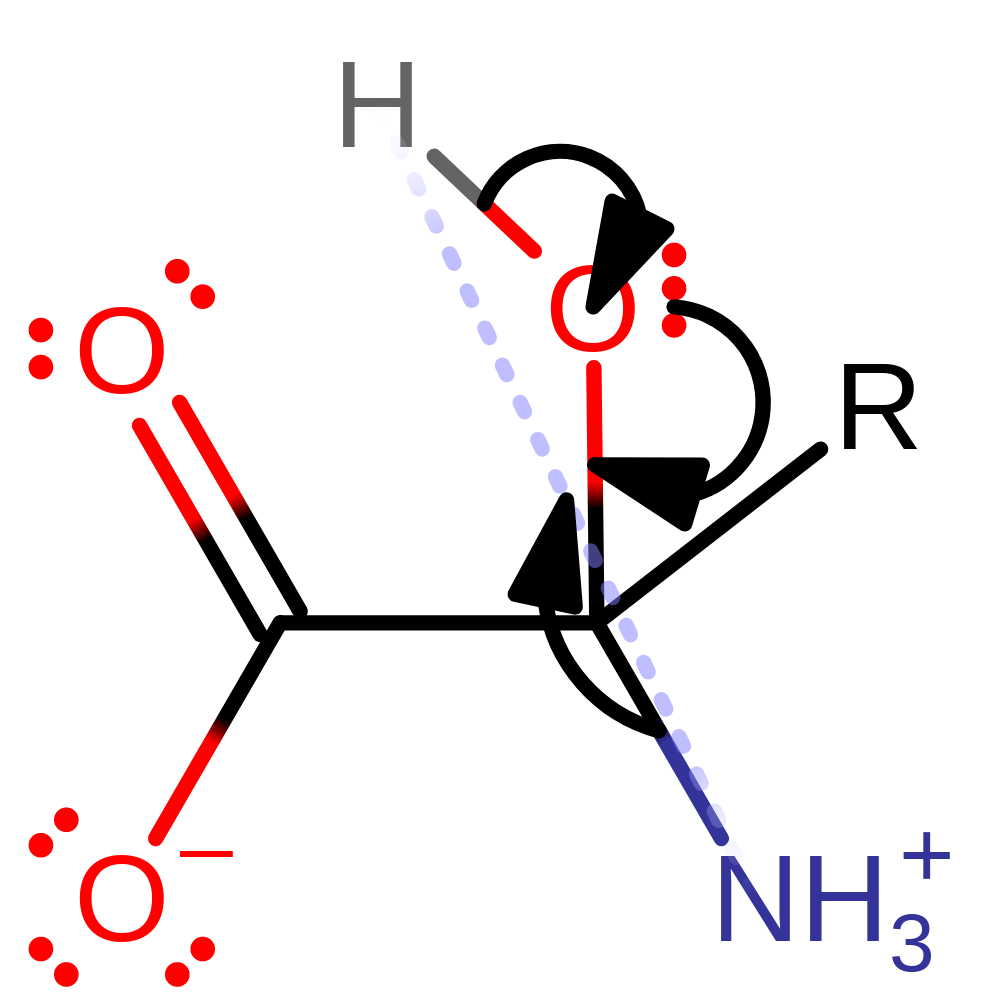
Step 6. Second half of the spontaneous hydrolysis of ammonia and the 2-oxo acid.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
deamination, reaction occurs outside the enzyme, ingold: intramolecular elimination, proton transferIntroduction
A carbanion mechanism has been proposed in which an enzyme base removes the alpha-proton (or possibly the amino proton) and so forms a carbanion intermediate. The resulting unstable carbanion would rapidly attack the N-5 locus of the flavin. Subsequent rapid rearrangement would result in reduced flavin and iminopyruvate. This mechanism was suggested upon the observation that pig kidney DAAO catalyses the elimination of halide from beta-halogenated amino acids.
Catalytic Residues Roles
| UniProt | PDB* (1c0p) | ||
| Ser335 (main-C), Gln339 (main-C), Asn54 (main-N), Asn54 (main-C) | Ser1335(337)A (main-C), Gln1339(341)A (main-C), Asn1054(56)A (main-N), Asn1054(56)A (main-C) | Act to lower the pKa of the alpha carbon proton. | modifies pKa |
| Ser335 | Ser1335(337)A | Acts as a general acid/base. | proton relay, proton acceptor, proton donor |
Chemical Components
proton transfer, hydride transfer, cofactor used, radical formation, electron transfer, overall reactant used, intermediate formation, radical termination, bimolecular homolytic addition, aromatic intramolecular elimination, native state of cofactor regenerated, intermediate terminated, overall product formed, native state of enzyme regenerated, bimolecular nucleophilic addition, reaction occurs outside the enzyme, intramolecular elimination, deaminationReferences
- Walsh CT et al. (1971), J Biol Chem, 246, 6855-6866. Studies on the mechanism of action of D-amino acid oxidase. Evidence for removal of substrate -hydrogen as a proton. PMID:4399475.
- Ghisla S et al. (2011), J Biol Chem, 286, 40987-40998. Revisitation of the Cl-Elimination Reaction of D-Amino Acid Oxidase: NEW INTERPRETATION OF THE REACTION THAT SPARKED FLAVOPROTEIN DEHYDROGENATION MECHANISMS. DOI:10.1074/jbc.m111.266536. PMID:21949129.
- Harris CM et al. (2001), Eur J Biochem, 268, 5504-5520. pH and kinetic isotope effects in d-amino acid oxidase catalysis. PMID:11683874.
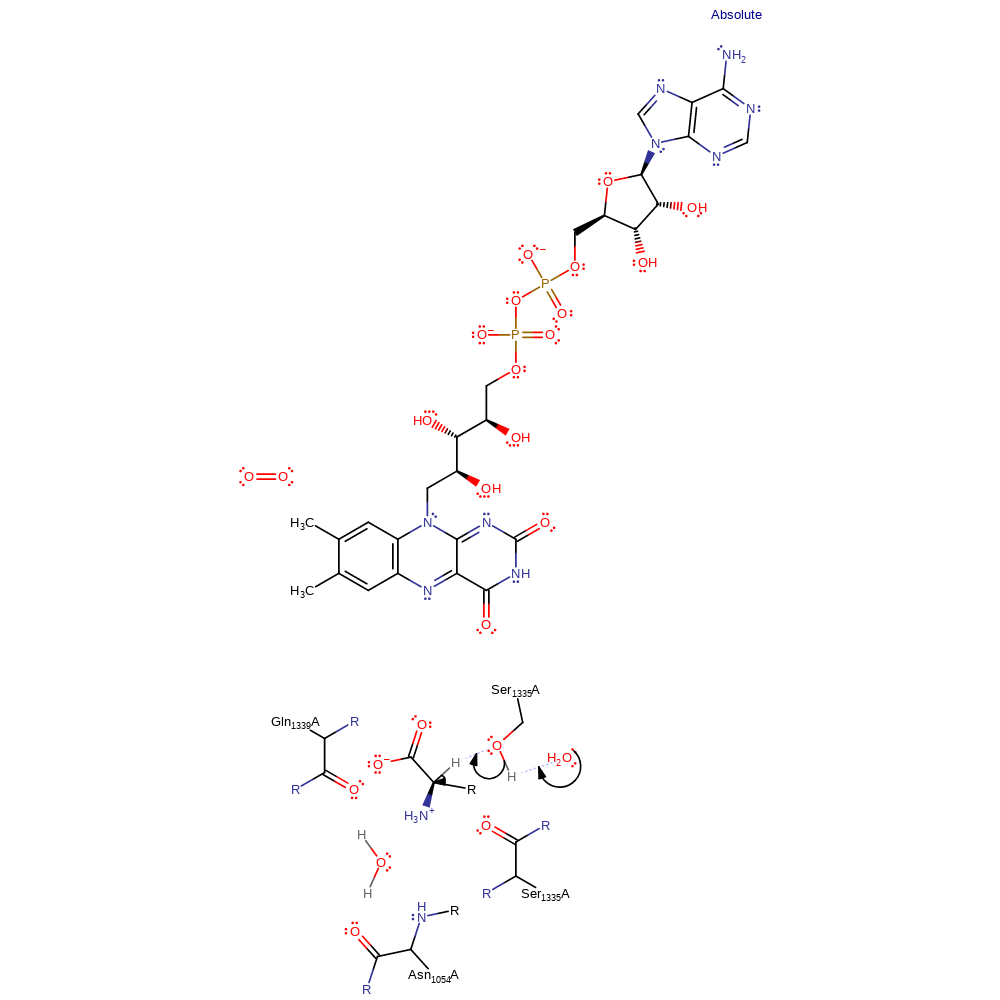
Step 1. Ser1335 abstracts a proton from the alpha carbon, forming a carbanion intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn1054(56)A (main-N) | modifies pKa |
| Asn1054(56)A (main-C) | modifies pKa |
| Ser1335(337)A (main-C) | modifies pKa |
| Gln1339(341)A (main-C) | modifies pKa |
| Ser1335(337)A | proton donor, proton relay, proton acceptor |
Chemical Components
proton transfer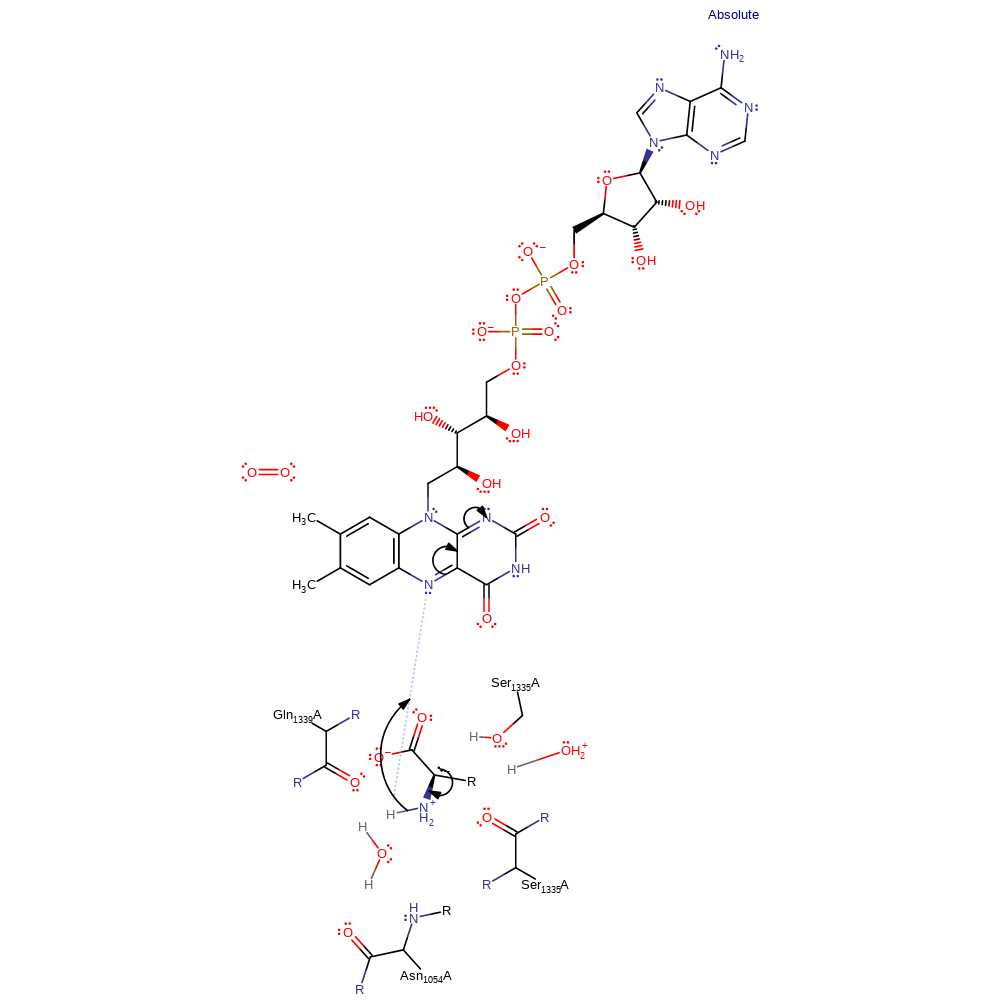
Step 2. The resulting unstable carbanion undergoes rearrangement to produce the reduced flavin and iminopyruvate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn1054(56)A (main-N) | modifies pKa |
| Asn1054(56)A (main-C) | modifies pKa |
| Ser1335(337)A (main-C) | modifies pKa |
| Gln1339(341)A (main-C) | modifies pKa |
Chemical Components
hydride transfer, cofactor used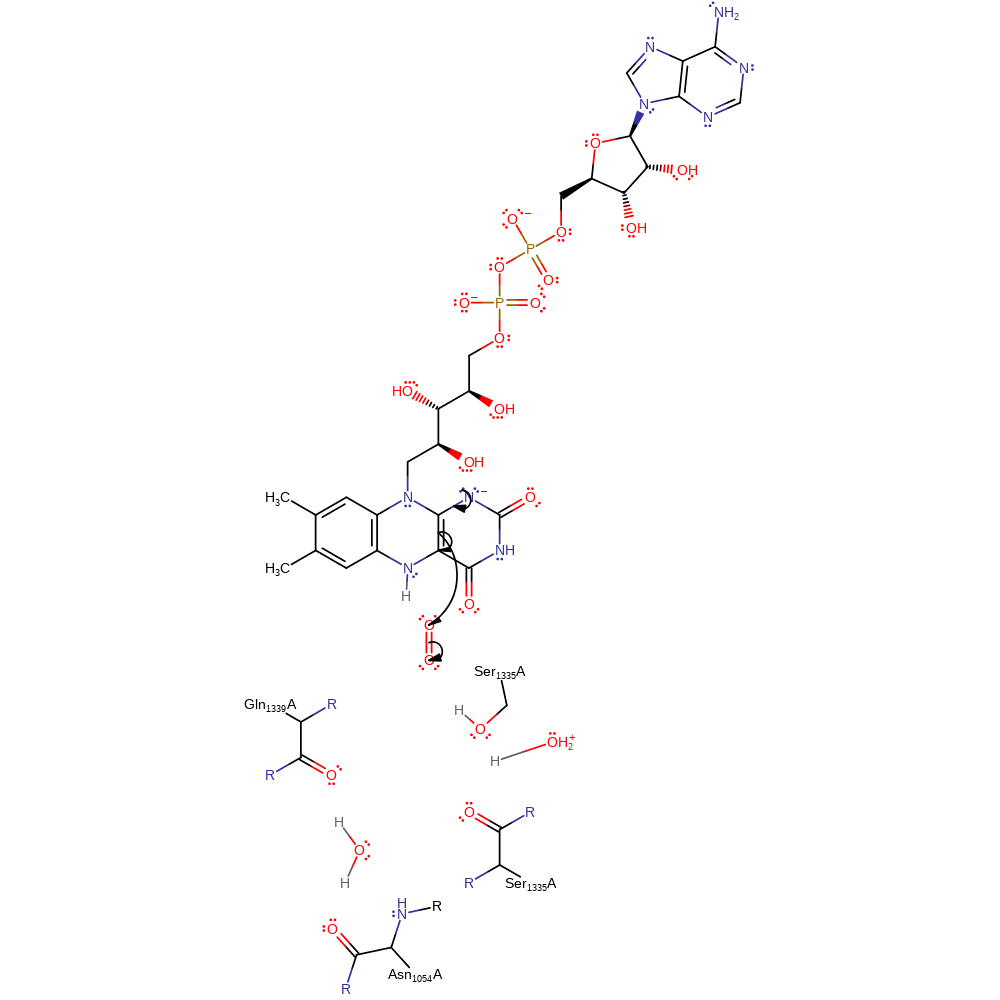
Step 3. The FAD undergoes double bond rearrangement which causes a single electron to be transferred to a dioxygen molecule.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
radical formation, electron transfer, overall reactant used, intermediate formation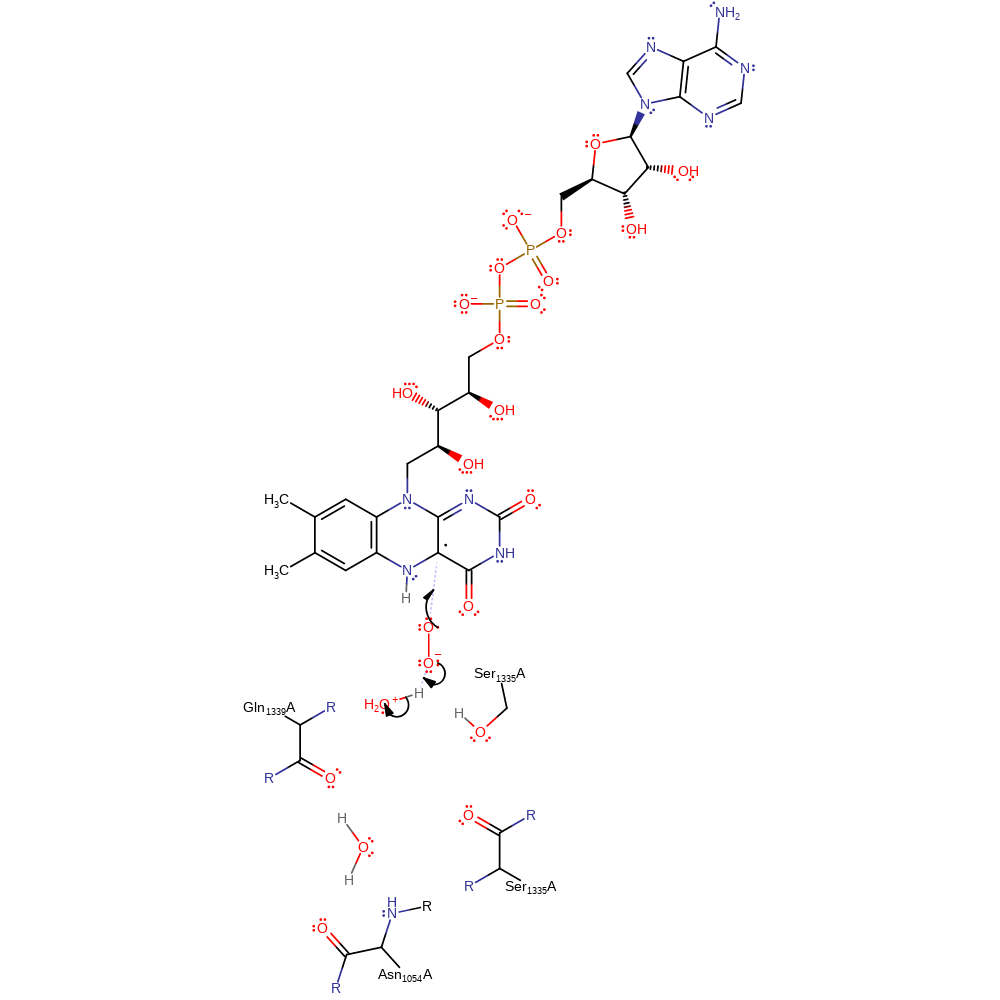
Step 4. The dioxygen molecule undergoes a homolytic reaction in which it colligates to FAD, with concomitant deprotonation of water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn1054(56)A (main-N) | hydrogen bond donor, hydrogen bond acceptor |
| Gln1339(341)A (main-C) | hydrogen bond acceptor |
Chemical Components
radical termination, ingold: bimolecular homolytic addition, proton transfer, intermediate formation
Step 5. The peroxo group deprotonates FAD, which initiates the elimination of hydrogen peroxide.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn1054(56)A (main-N) | hydrogen bond donor, hydrogen bond acceptor |
| Gln1339(341)A (main-C) | hydrogen bond acceptor |
Chemical Components
ingold: aromatic intramolecular elimination, native state of cofactor regenerated, intermediate terminated, overall product formed, native state of enzyme regenerated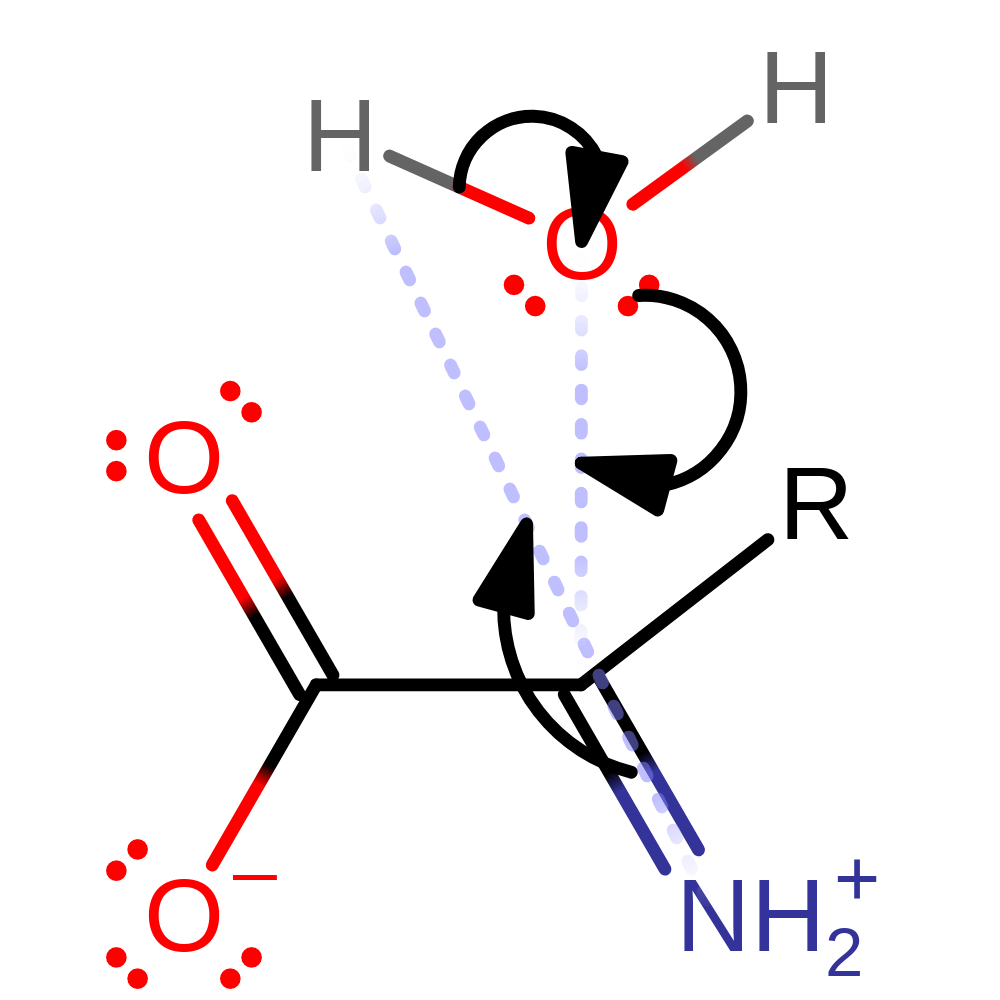
Step 6. The product of the enzyme undergoes spontaneous hydrolysis outside of the active site to produce ammonium and the 2-oxo acid.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, reaction occurs outside the enzyme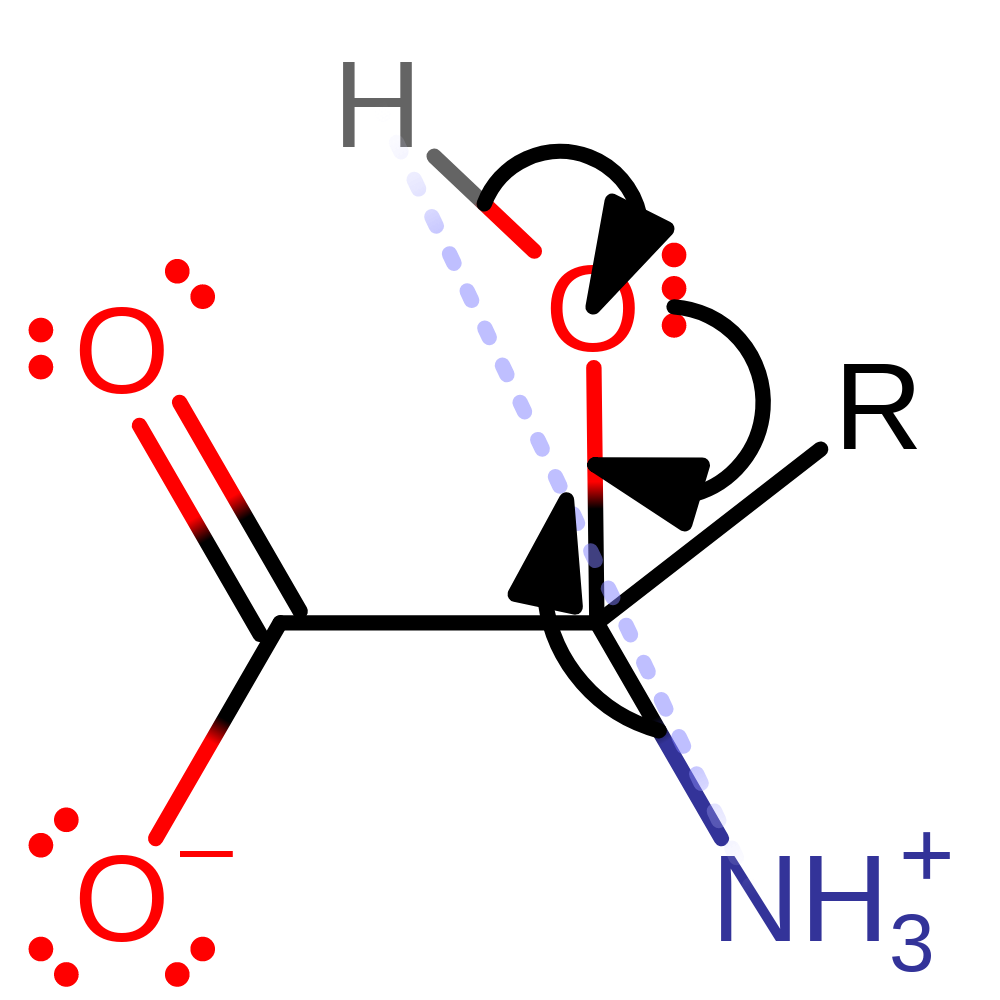
Step 7. Second half of the spontaneous hydrolysis of ammonia and the 2-oxo acid.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|






 Download:
Download: 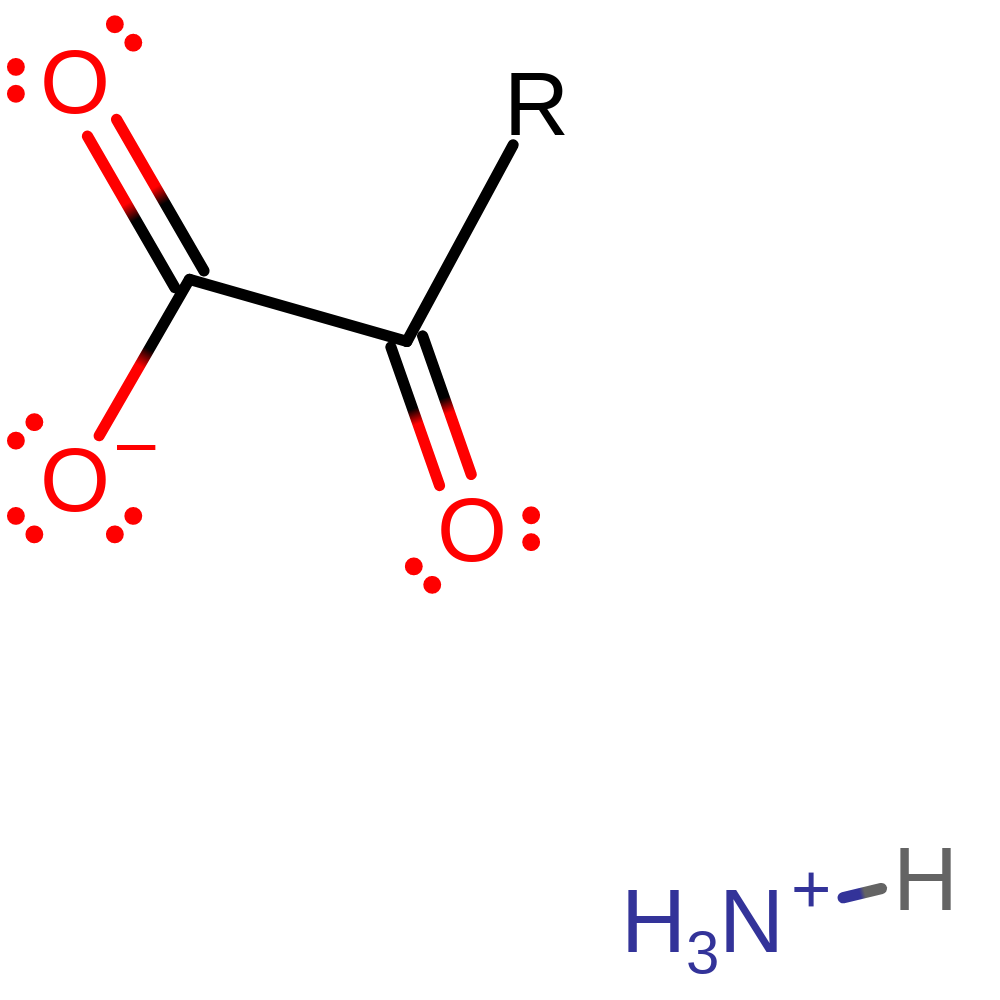 Download:
Download: