Creatinase
Creatinase or creatine amidinohydrolase (EC:3.5.3.3) catalyses the conversion of creatine and water to sarcosine and urea. This is a key step in the metabolic breakdown of creatinine by micro-organisms. Creatinase is also found in higher animals but its metabolic role is as yet not known.
The enzyme works as a homodimer, and is induced by choline chloride. Each monomer of creatinase has two clearly defined domains, a small N-terminal domain, and a large C-terminal domain. The C-terminal domain is a member of the MEROPS peptidase family M24 (clan MG), which share a common structural-fold, the "pita-bread" fold. The active site is located between the N- and C-terminal domains.
Reference Protein and Structure
- Sequence
-
P38488
 (3.5.3.3)
(3.5.3.3)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Pseudomonas putida (Bacteria)

- PDB
-
1chm
- ENZYMATIC MECHANISM OF CREATINE AMIDINOHYDROLASE AS DEDUCED FROM CRYSTAL STRUCTURES
(1.9 Å)



- Catalytic CATH Domains
-
3.90.230.10
 (see all for 1chm)
(see all for 1chm)
- Cofactors
- Water (1)
Enzyme Mechanism
Introduction
Histidine 232 abstracts a proton from an active site water molecule to create a hydroxide nucleophile. Glutamate 262 and 358 are negatively charged and break the resonance of the guanidinium group, this allows the hydroxide to attack at C1. Histidine 232 now donates its proton to N3 which leads to the C1-N3 bond breaking. Histidine 232 again abstracts a proton, this time from the hydroxide attached to C1, leaving the reaction products urea and sarcosine.
Catalytic Residues Roles
| UniProt | PDB* (1chm) | ||
| His232 | His232(231)A | The residue acts as a general base to the hydrolytic water molecule, a general acid to the intermediate, and then as a base once more in a heterolytic bimolecular elimination reaction, leading to the products. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Glu358, Glu262 | Glu358(357)A, Glu262(261)A | The residue's charged side chain breaks the resonance of the creatine gaunidinium group, allowing the hydroxyl to act as a nucleophile at the partial carbocation. | hydrogen bond acceptor, electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic addition, intermediate formation, bimolecular elimination, intermediate collapse, intermediate terminated, native state of enzyme regenerated, inferred reaction stepReferences
- Coll M et al. (1990), J Mol Biol, 214, 597-610. Enzymatic mechanism of creatine amidinohydrolase as deduced from crystal structures. DOI:10.1016/0022-2836(90)90201-v. PMID:1696320.
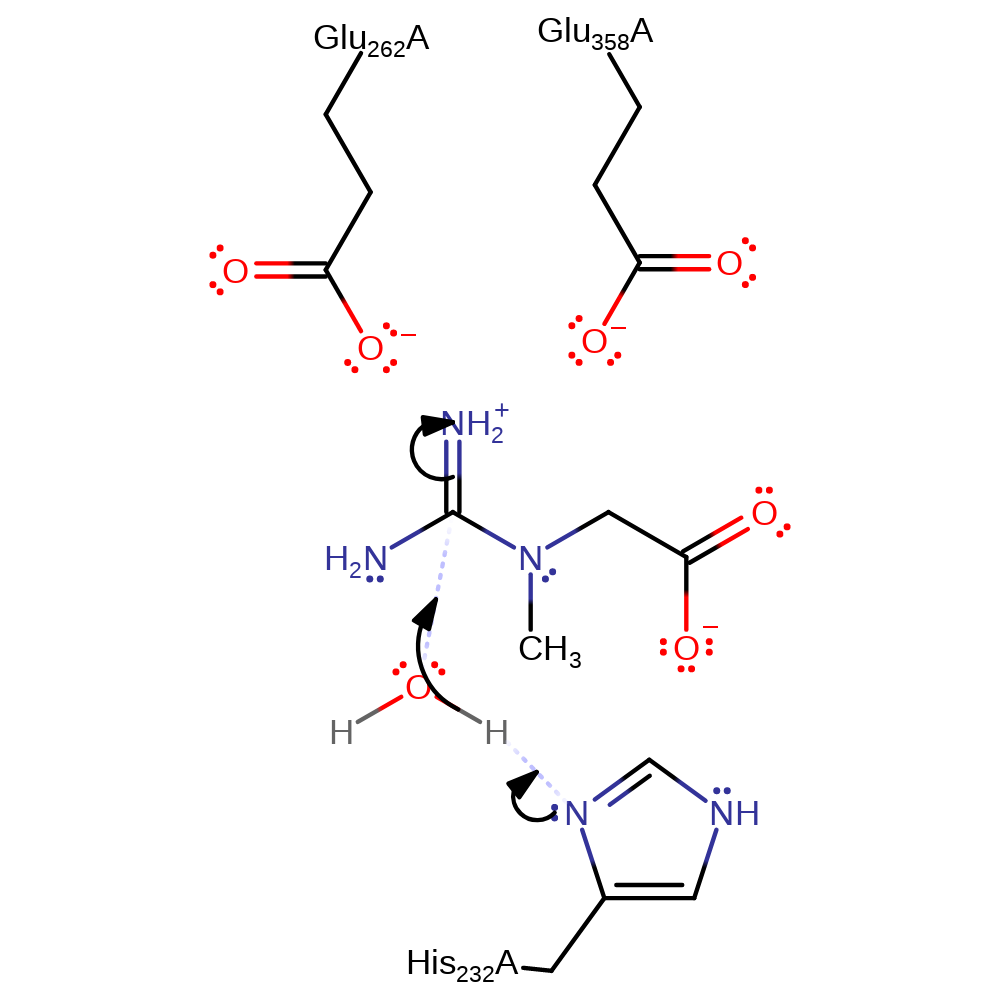
Step 1. His232 deprotonates water, which initiates a nucleophilic attack on the guanidinium carbon of the substrate in an addition reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu358(357)A | hydrogen bond acceptor, electrostatic stabiliser |
| His232(231)A | hydrogen bond acceptor |
| Glu262(261)A | hydrogen bond acceptor, electrostatic stabiliser |
| His232(231)A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation
Step 2. The methylated nitrogen of the guanidinium group deprotonates His232.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu358(357)A | hydrogen bond acceptor |
| His232(231)A | hydrogen bond donor |
| Glu262(261)A | hydrogen bond acceptor |
| His232(231)A | proton donor |
Chemical Components
proton transfer, intermediate formation
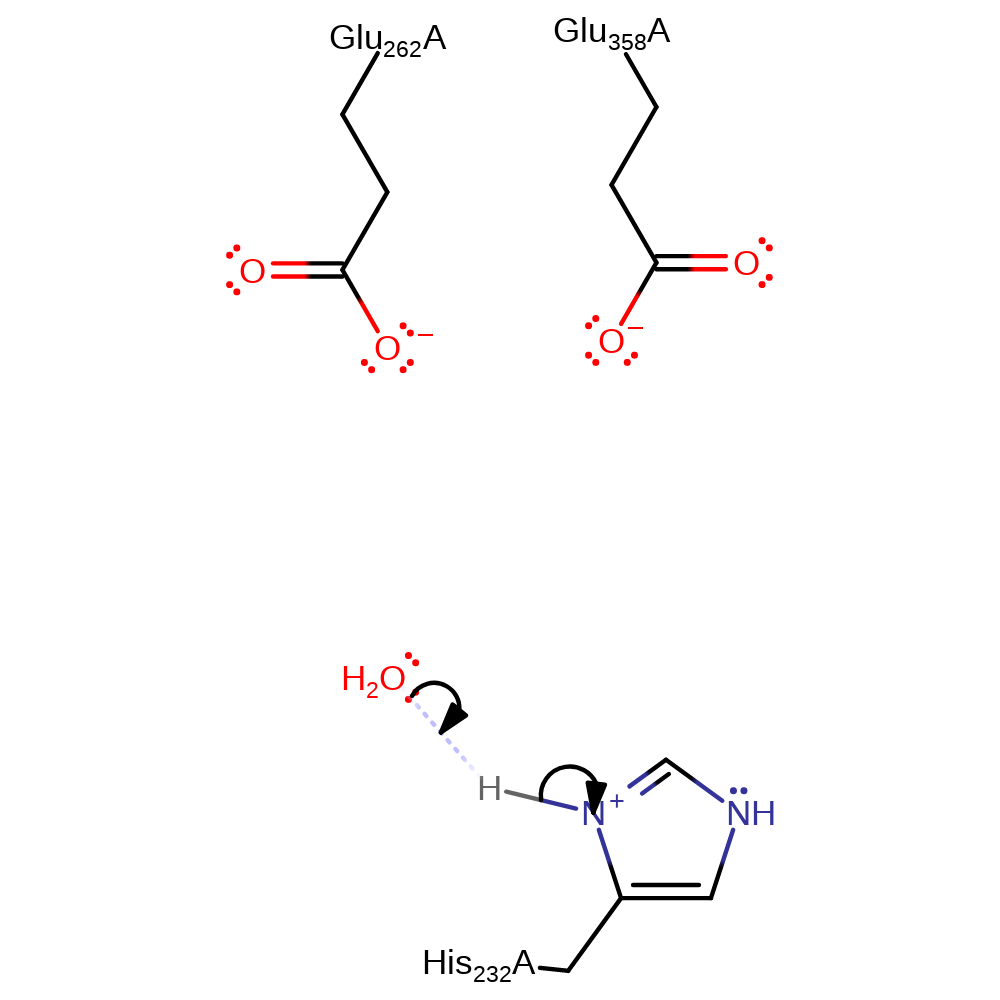
Step 3. His232 deprotonates the hydroxyl group added, eliminating sarcosine.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu358(357)A | hydrogen bond acceptor |
| His232(231)A | hydrogen bond acceptor |
| Glu262(261)A | hydrogen bond acceptor |
| His232(231)A | proton acceptor |
Chemical Components
ingold: bimolecular elimination, intermediate collapse, intermediate terminatedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His232(231)A | hydrogen bond donor, proton donor |




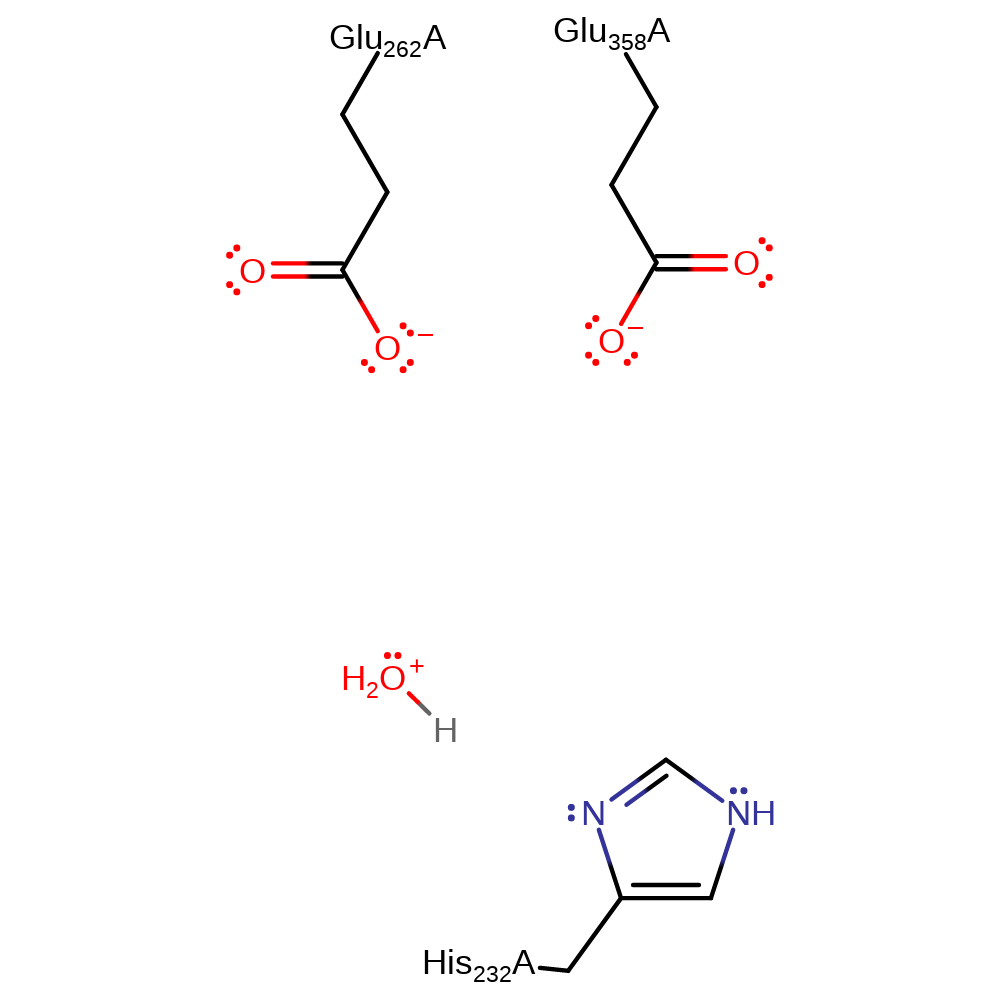 Download:
Download: