Aspartate-ammonia ligase
L-asparagine synthetase in Escherichia coli converts L-aspartate and ammonia to L-asparagine, with the simultaneous hydrolysis of ATP to AMP and diphosphate. The reaction involves the formation of an aminoacyl-adenylate intermediate.
Reference Protein and Structure
- Sequence
-
P00963
 (6.3.1.1)
(6.3.1.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
12as
- ASPARAGINE SYNTHETASE MUTANT C51A, C315A COMPLEXED WITH L-ASPARAGINE AND AMP
(2.2 Å)



- Catalytic CATH Domains
-
3.30.930.10
 (see all for 12as)
(see all for 12as)
- Cofactors
- Magnesium(2+) (1) Metal MACiE
Enzyme Reaction (EC:6.3.1.1)
Enzyme Mechanism
Introduction
The carboxylate group of the L-aspartate side chain acts as a nucleophile and attacks the alpha-phosphate of ATP in a substitution reaction that liberates pyrophosphate. Arg100 and Gln116 stabilise the intermediates formed. Asp46 deprotonates the ammonia molecule, which acts as a nucleophile to attack the carbonyl group (attached to the phosphate) of the L-aspartate in an addition reaction. Arg100 and Gln116 stabilise the intermediates formed. The tetrahedral intermediate collapses to reform the carbonyl, cleaving the P-O bond and liberating AMP and the L-asparagine. Asp46 donates its proton back to the free AMP. Arg100 and Gln116 stabilise the intermediates formed.
An Mg(II) ion is also believed to be involved in catalysis, stabilising negative charges along with Arg100 and Gln116 in each transition state or intermediate, but this is not present in the crystal structure.
Catalytic Residues Roles
| UniProt | PDB* (12as) | ||
| Arg100 | Arg100A | Forms part of oxyanion hole to stabilise the negative charge on substrate oxygen during the transition state. | hydrogen bond donor, electrostatic stabiliser |
| Asp235, Glu248 | Asp235A, Glu248A | Forms part of the magnesium binding site. | metal ligand |
| Asp46 | Asp46A | Deprotonates the ammonia molecule to activate it as a nucleophile, and donates the proton back to AMP. Also stabilises the positive charge on the substrate nitrogen during the transition state. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Gln116 | Gln116A | The NH2 group forms part of oxyanion hole to stabilise the negative charge on oxygen during the transition state. The oxygen of Gln116 also stabilises the positive charge on the substrate nitrogen during the transition state. | hydrogen bond donor, electrostatic stabiliser |
| Ser251, Ser251 (main-C) | Ser251A, Ser251A (main-C) | Helps position and stabilise the intermediates during the course of the reaction. | electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic substitution, overall reactant used, overall product formed, dephosphorylation, intermediate formation, proton transfer, bimolecular nucleophilic addition, unimolecular elimination by the conjugate base, intermediate collapse, intermediate terminated, native state of enzyme regeneratedReferences
- Koizumi M et al. (1999), J Am Chem Soc, 121, 5799-5800. A Potent Transition-State Analogue Inhibitor ofEscherichia coliAsparagine Synthetase A. DOI:10.1021/ja990851a.
- Manhas R et al. (2014), J Biol Chem, 289, 12096-12108. Identification and Functional Characterization of a Novel Bacterial Type Asparagine Synthetase A: A tRNA SYNTHETASE PARALOG FROM LEISHMANIA DONOVANI. DOI:10.1074/jbc.m114.554642. PMID:24610810.
- Kolodkin-Gal I et al. (2007), Science, 318, 652-655. A Linear Pentapeptide Is a Quorum-Sensing Factor Required for mazEF-Mediated Cell Death in Escherichia coli. DOI:10.1126/science.1147248. PMID:17962566.
- Nakatsu T et al. (1998), Nat Struct Biol, 5, 15-19. Crystal structure of asparagine synthetase reveals a close evolutionary relationship to class II aminoacyl-tRNA synthetase. DOI:10.1038/nsb0198-15. PMID:9437423.
- Hinchman SK et al. (1992), J Biol Chem, 267, 144-149. A relationship between asparagine synthetase A and aspartyl tRNA synthetase. PMID:1346128.
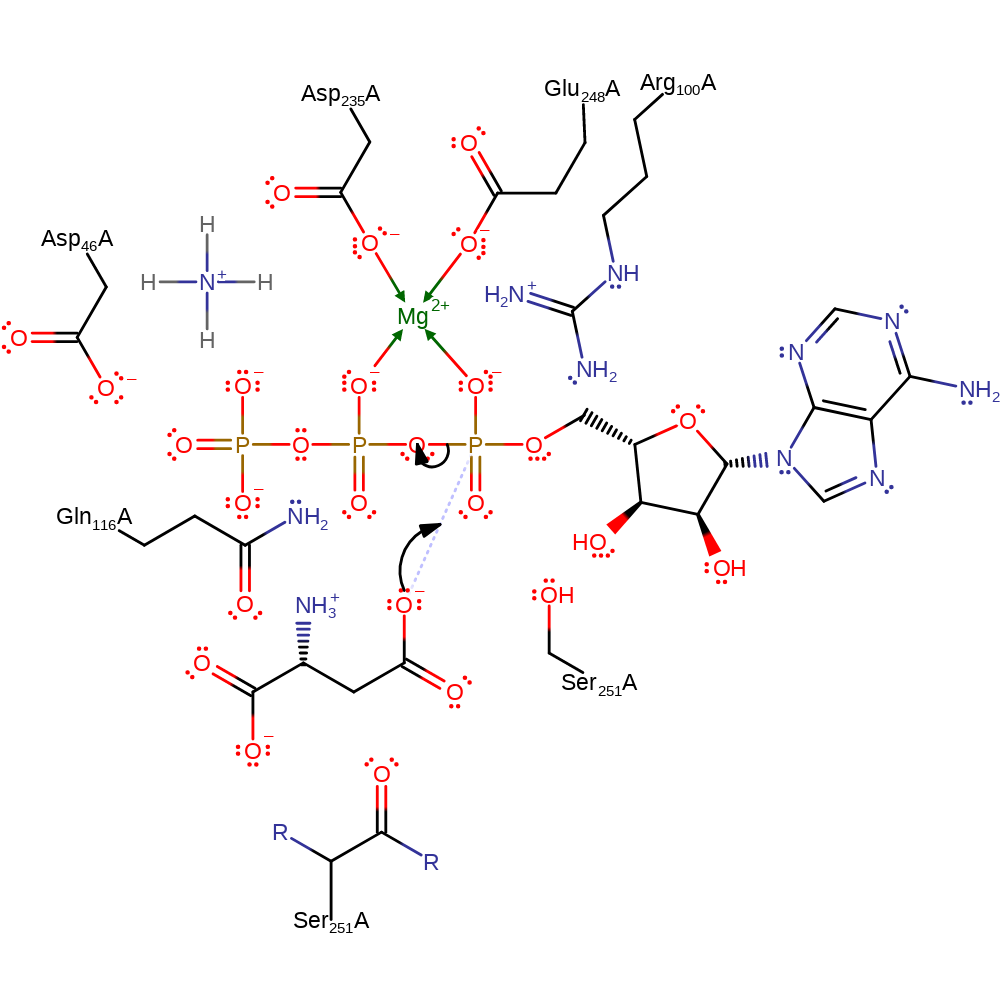
Step 1. The carboxylate group of the L-aspartate side chain acts as a nucleophile and attacks the alpha-phosphate of ATP in a substitution reaction that liberates pyrophosphate. Mg(II), Arg100 and Gln116 stabilise the intermediates formed.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg100A | hydrogen bond donor, electrostatic stabiliser |
| Gln116A | hydrogen bond donor, electrostatic stabiliser |
| Ser251A | electrostatic stabiliser |
| Ser251A (main-C) | electrostatic stabiliser |
| Glu248A | metal ligand |
| Asp235A | metal ligand |
Chemical Components
ingold: bimolecular nucleophilic substitution, overall reactant used, overall product formed, dephosphorylation, intermediate formation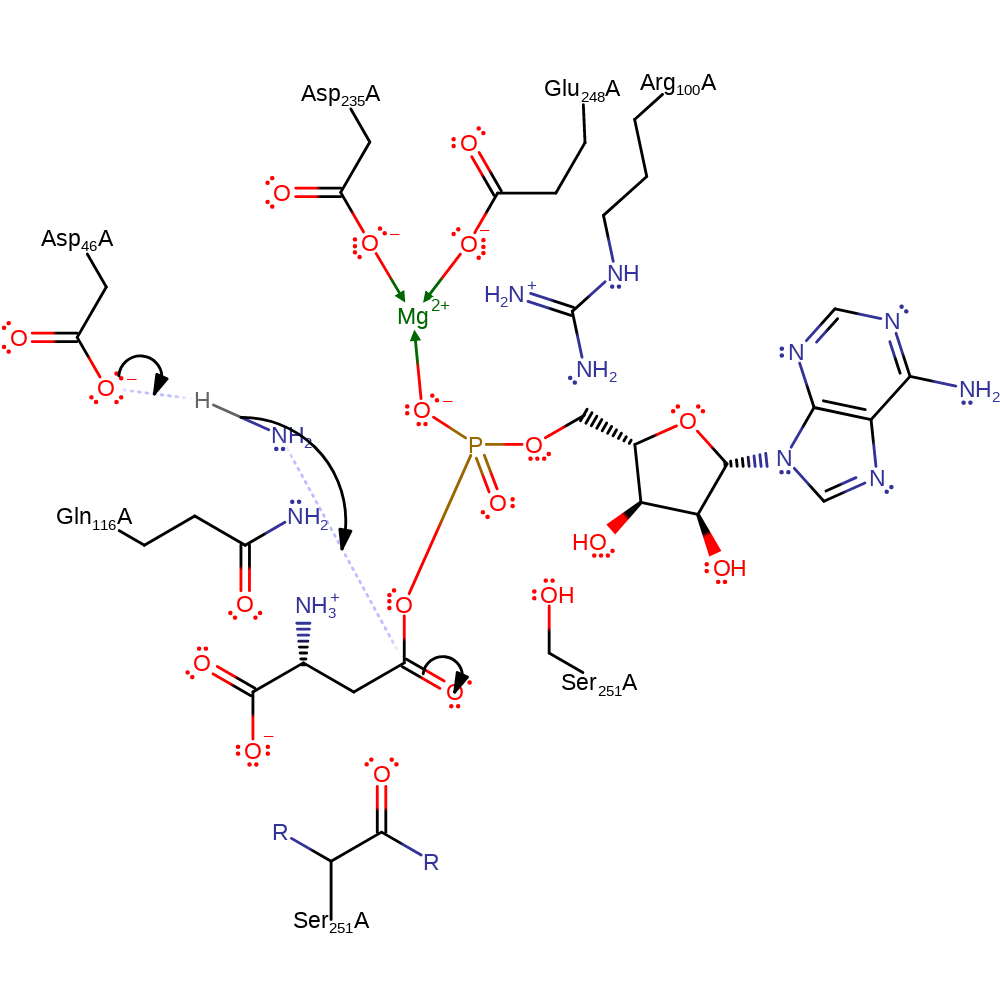
Step 2. Asp46 deprotonates the ammonia molecule, which acts as a nucleophile to attack the carbonyl group (attached to the phosphate) of the L-aspartate in an addition reaction. Mg(II), Arg100 and Gln116 stabilise the intermediates formed.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp46A | hydrogen bond acceptor |
| Arg100A | hydrogen bond donor, electrostatic stabiliser |
| Gln116A | hydrogen bond donor, electrostatic stabiliser |
| Ser251A | electrostatic stabiliser |
| Ser251A (main-C) | electrostatic stabiliser |
| Glu248A | metal ligand |
| Asp235A | metal ligand |
| Asp46A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, overall reactant used, intermediate formation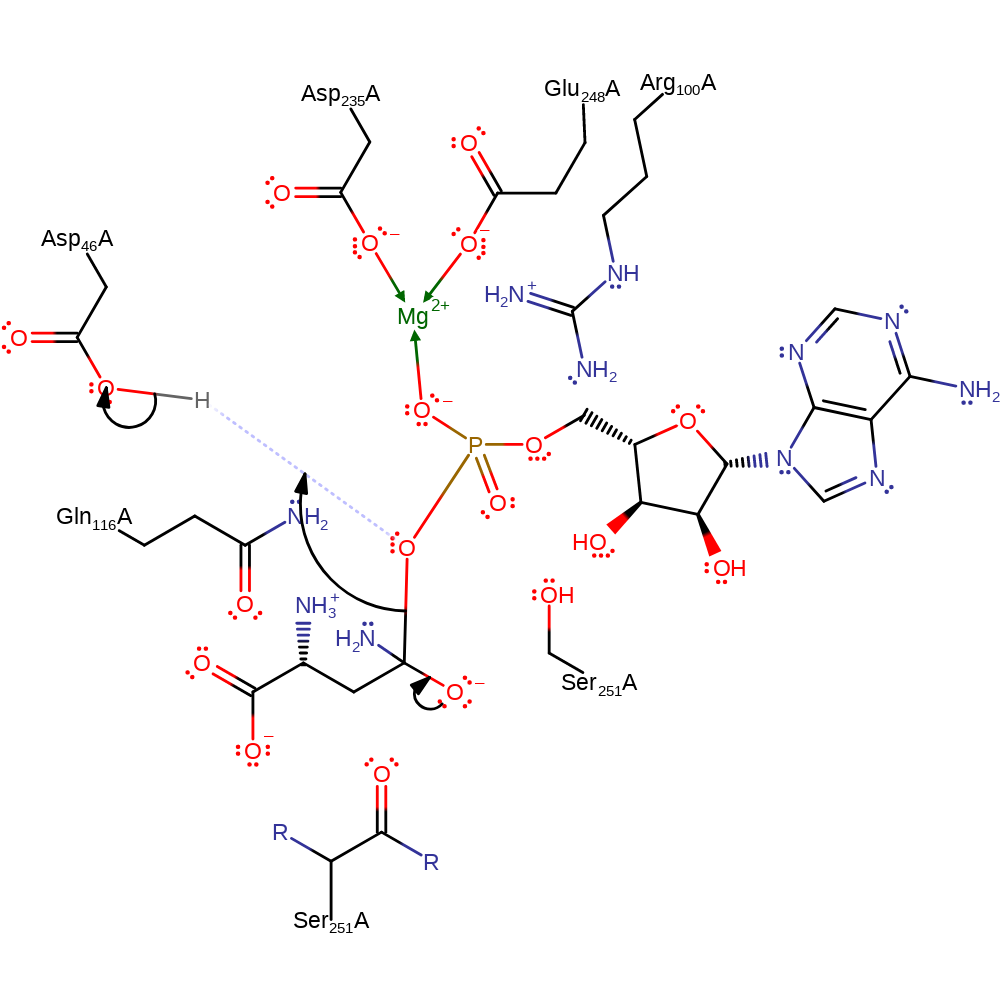
Step 3. The oxyanion formed re-forms the carbonyl group, cleaving the P-O bond in a conjugate base elimination reaction liberating AMP and the L-asparagine. Asp46 donates its proton back to the liberated AMP. Mg(II), Arg100 and Gln116 stabilise the intermediates formed.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln116A | hydrogen bond donor, electrostatic stabiliser |
| Asp46A | hydrogen bond donor |
| Arg100A | hydrogen bond donor, electrostatic stabiliser |
| Glu248A | metal ligand |
| Asp235A | metal ligand |
| Ser251A | electrostatic stabiliser |
| Ser251A (main-C) | electrostatic stabiliser |
| Asp46A | proton donor |

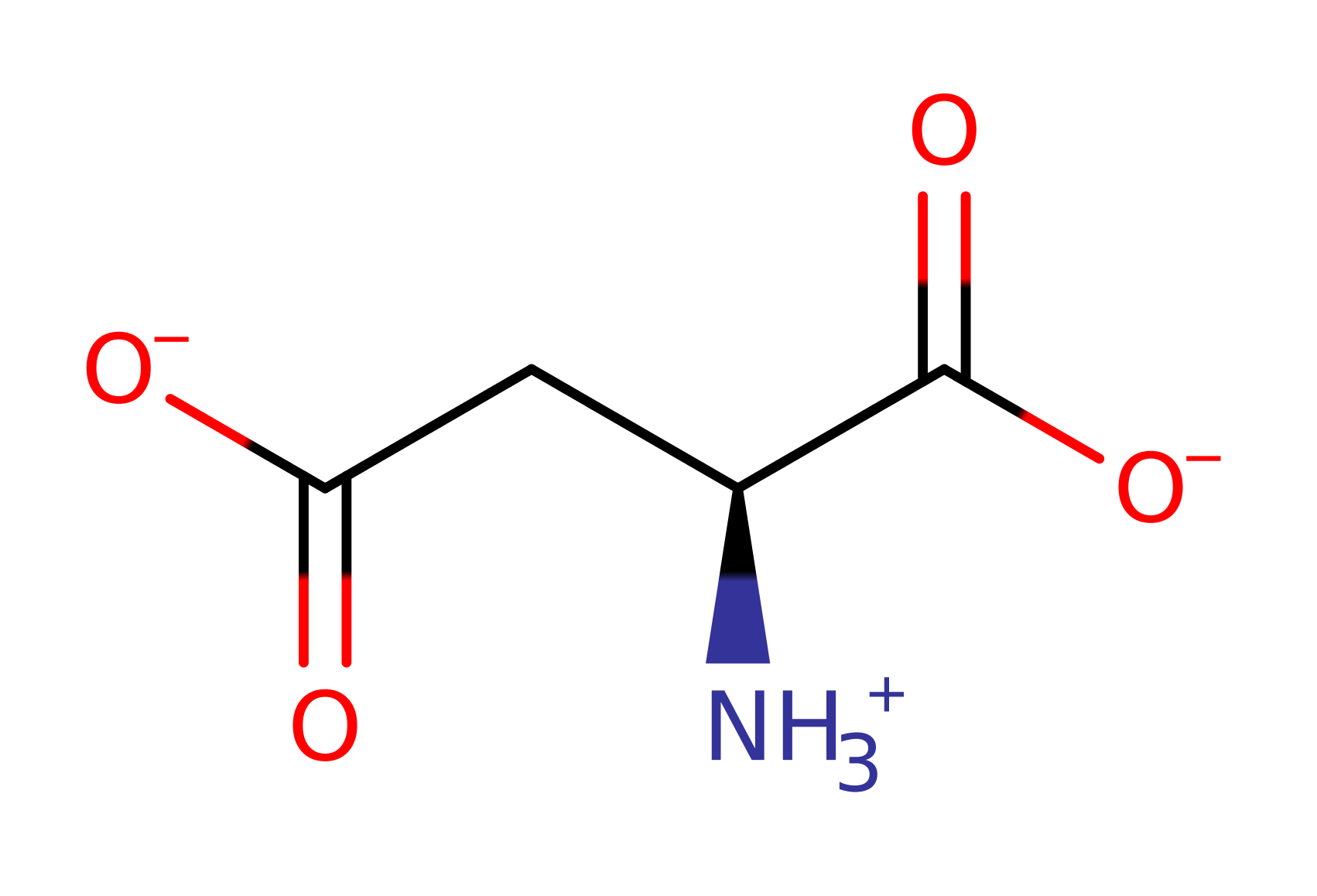




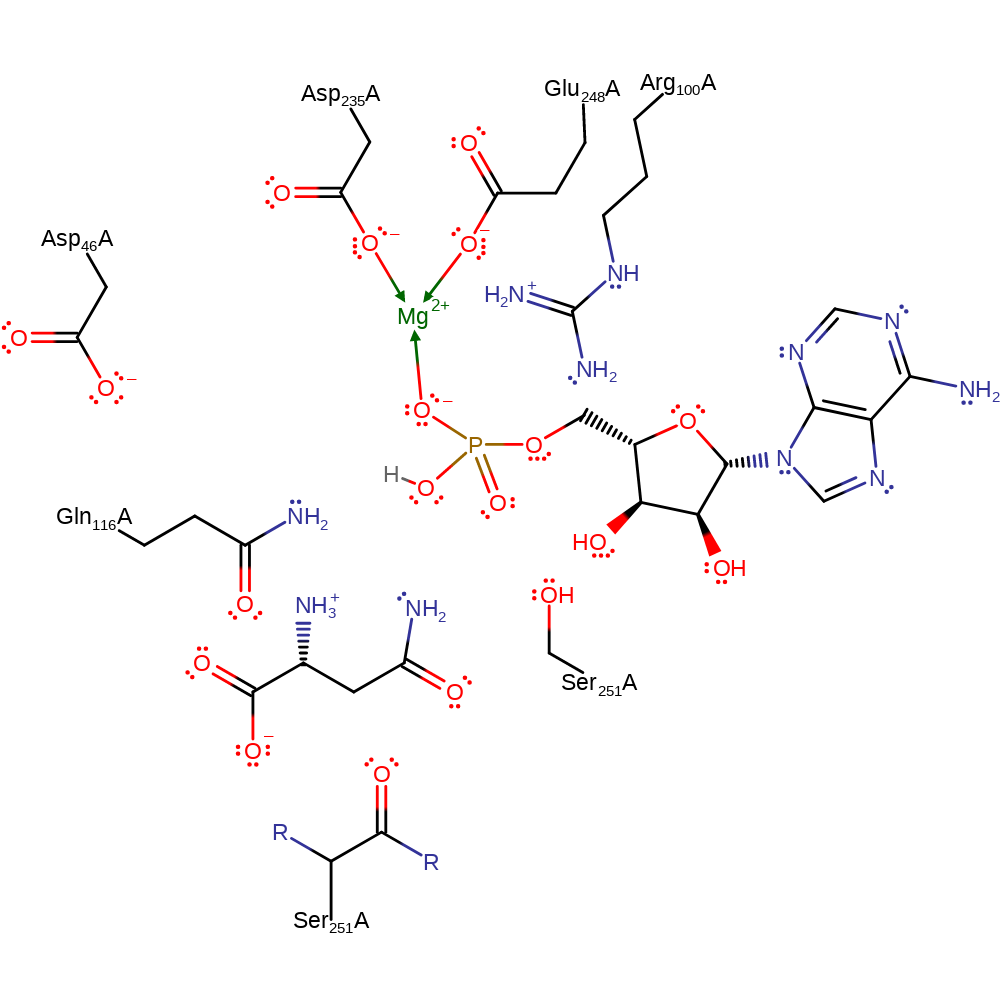 Download:
Download: