Phosphoenolpyruvate carboxykinase (ATP)
Phosphoenolpyruvate carboxykinase (PEPCK) catalyses first committed (rate-limiting) step in hepatic gluconeogenesis, namely the reversible decarboxylation of oxaloacetate to phosphoenolpyruvate (PEP) and carbon dioxide. This reaction can occur using either ATP or GTP as a source of the phosphate. The ATP-utilising (EC:4.1.1.49, represented here) and GTP-utilising (EC:4.1.1.32) enzymes form two divergent subfamilies, which have little sequence similarity but which retain conserved active site residues.
The reaction requires two divalent cations for activity, usually magnesium and manganese. One cation interacts with the enzyme at metal binding site 1 to elicit activation, while the second cation interacts at metal binding site 2 to serve as a metal-nucleotide substrate. In bacteria, fungi and plants, PEPCK is involved in the glyoxylate bypass, an alternative to the tricarboxylic acid cycle.
PEPCK consists of an N-terminal and a catalytic C-terminal domain, with the active site and metal ions located in a cleft between them. Both domains have an alpha/beta topology that is partly similar to one another [PMID:15023367, PMID:8609605]. Substrate binding causes PEPCK to undergo a conformational change, which accelerates catalysis by forcing bulk solvent molecules out of the active site [PMID:15890557]. PCK uses an alpha/beta/alpha motif for nucleotide binding, this motif differing from other kinase domains.
Reference Protein and Structure
- Sequence
-
P22259
 (4.1.1.49)
(4.1.1.49)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1aq2
- PHOSPHOENOLPYRUVATE CARBOXYKINASE
(1.9 Å)



- Catalytic CATH Domains
-
2.170.8.10
 3.40.449.10
3.40.449.10  3.90.228.20
3.90.228.20  (see all for 1aq2)
(see all for 1aq2)
- Cofactors
- Magnesium(2+) (1), Manganese(2+) (1) Metal MACiE
Enzyme Reaction (EC:4.1.1.49)
Enzyme Mechanism
Introduction
The substrate undergoes decarboxylation, resulting in the formation of an enol intermediate, stabilised by positively charged residues. The oxyanion formed attacks the gamma phosphate of ATP in a nucleophilic substitution, giving the products of ADP and phosphoenolpyruvate.
Catalytic Residues Roles
| UniProt | PDB* (1aq2) | ||
| Arg333 | Arg333A | Activates substrate, stabilises enolate intermediate. | hydrogen bond donor, electrostatic stabiliser |
| His232 | His232A | Stabilises intermediates. | hydrogen bond donor, metal ligand, electrostatic stabiliser |
| Lys254 | Lys254A | Neutralises repulsions between phosphate oxygen atoms on ATP, promoting the formation of a high energy conformer. | hydrogen bond donor, electrostatic stabiliser |
| Asp269, His232, Lys213 | Asp269A, His232A, Lys213A | Forms part of the Mn(II) binding site, along with the gamma phosphate of ATP. | metal ligand |
| Ser250 | Ser250A | The hydroxyl group of serine 250 (252 in the yeast protein) is required for proper oxaloacetate interaction. | steric role |
| Lys213, Arg65 | Lys213A, Arg65A | Forms part of the carbon dioxide binding site. In the reaction direction shown in this entry (decarboxylation) it is responsible for stabilising the transition states and binding. In the reverse direction it is responsible for binding and increasing the electrophilicity of the carbon dioxide. | metal ligand |
| Thr255 | Thr255A | Forms part of the Mg(II) binding site, along with beta and gamma phosphates of ATPO. | metal ligand |
Chemical Components
unimolecular elimination by the conjugate base, decarboxylation, bimolecular nucleophilic substitutionReferences
- Matte A et al. (1997), J Biol Chem, 272, 8105-8108. Structure and Mechanism of Phosphoenolpyruvate Carboxykinase. DOI:10.1074/jbc.272.13.8105. PMID:9139042.
- Johnson TA et al. (2012), Biochemistry, 51, 9547-9559. The Ω-Loop Lid Domain of Phosphoenolpyruvate Carboxykinase Is Essential for Catalytic Function. DOI:10.1021/bi301278t. PMID:23127136.
- Pérez E et al. (2010), Protein J, 29, 299-305. Saccharomyces cerevisiae Phosphoenolpyruvate Carboxykinase: The Relevance of Glu299 and Leu460 for Nucleotide Binding. DOI:10.1007/s10930-010-9252-6. PMID:20524049.
- Sepúlveda C et al. (2010), Biochimie, 92, 814-819. Electrostatic interactions play a significant role in the affinity of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase for Mn2+. DOI:10.1016/j.biochi.2010.02.032. PMID:20211682.
- Castillo D et al. (2009), Biochimie, 91, 295-299. Functional evaluation of serine 252 of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase. DOI:10.1016/j.biochi.2008.10.005. PMID:18996167.
- Yévenes A et al. (2007), Protein J, 26, 135-141. Relevance of phenylalanine 216 in the affinity of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase for Mn(II). DOI:10.1007/s10930-006-9054-z. PMID:17195942.
- Cotelesage JJ et al. (2007), Int J Biochem Cell Biol, 39, 1204-1210. How does an enzyme recognize CO2? DOI:10.1016/j.biocel.2007.03.015. PMID:17475535.
- Yévenes A et al. (2006), Biochimie, 88, 663-672. Site-directed mutagenesis study of the microenvironment characteristics of Lys213 of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase. DOI:10.1016/j.biochi.2005.12.002. PMID:16469427.
- Villarreal JM et al. (2006), Int J Biochem Cell Biol, 38, 576-588. Nucleotide specificity of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinaseKinetics, fluorescence spectroscopy, and molecular simulation studies. DOI:10.1016/j.biocel.2005.10.018. PMID:16330239.
- Jabalquinto AM et al. (2004), Biochimie, 86, 47-51. Anaerobiospirillum succiniciproducens phosphoenolpyruvate carboxykinase. Mutagenesis at metal site 1. DOI:10.1016/j.biochi.2003.10.013. PMID:14987800.
- Cristina Ravanal M et al. (2004), Biochimie, 86, 357-362. Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase: relevance of arginine 70 for catalysis. DOI:10.1016/j.biochi.2004.06.001. PMID:15358051.
- Sudom A et al. (2003), J Bacteriol, 185, 4233-4242. Mechanisms of activation of phosphoenolpyruvate carboxykinase from Escherichia coli by Ca2+ and of desensitization by trypsin. PMID:12837799.
- González-Nilo FD et al. (2002), Biochim Biophys Acta Proteins Proteomics, 1599, 65-71. Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase: theoretical and experimental study of the effect of glutamic acid 284 on the protonation state of lysine 213. DOI:10.1016/s1570-9639(02)00400-4.
- Llanos L et al. (2001), FEBS Lett, 493, 1-5. Mutation Arg336 to Lys inSaccharomyces cerevisiaephosphoenolpyruvate carboxykinase originates an enzyme with increased oxaloacetate decarboxylase activity. DOI:10.1016/s0014-5793(01)02158-5.
- Sudom AM et al. (2001), J Mol Biol, 314, 83-92. The phosphoryl-transfer mechanism of Escherichia coli phosphoenolpyruvate carboxykinase from the use of AlF3. DOI:10.1006/jmbi.2001.5120. PMID:11724534.
- Bazaes S et al. (1997), Biochim Biophys Acta, 1337, 166-174. Identification of reactive conserved histidines in phosphoenolpyruvate carboxykinases from Escherichia coli and Saccharomyces cerevisiae. DOI:10.1016/s0167-4838(96)00155-0. PMID:9048893.
- Tari LW et al. (1997), Nat Struct Biol, 4, 990-994. Mg2+–Mn2+ clusters in enzyme-catalyzed phosphoryl-transfer reactions. DOI:10.1038/nsb1297-990. PMID:9406547.
- Tari LW et al. (1996), Nat Struct Biol, 3, 355-363. Snapshot of an enzyme reaction intermediate in the structure of the ATP–Mg2+–oxalate ternary complex of Escherichia coli PEP carboxykinase. DOI:10.1038/nsb0496-355. PMID:8599762.
- Matte A et al. (1996), J Mol Biol, 256, 126-143. Crystal Structure ofEscherichia coliPhosphoenolpyruvate Carboxykinase: A New Structural Family with the P-loop Nucleoside Triphosphate Hydrolase Fold. DOI:10.1006/jmbi.1996.0072. PMID:8609605.
- Medina V et al. (1990), J Bacteriol, 172, 7151-7156. Sequence of the pckA gene of Escherichia coli K-12: relevance to genetic and allosteric regulation and homology of E. coli phosphoenolpyruvate carboxykinase with the enzymes from Trypanosoma brucei and Saccharomyces cerevisiae. PMID:1701430.
- Goldie AH et al. (1980), J Biol Chem, 255, 1399-1405. Allosteric control by calcium and mechanism of desensitization of phosphoenolpyruvate carboxykinase of Escherichia coli. PMID:6986370.
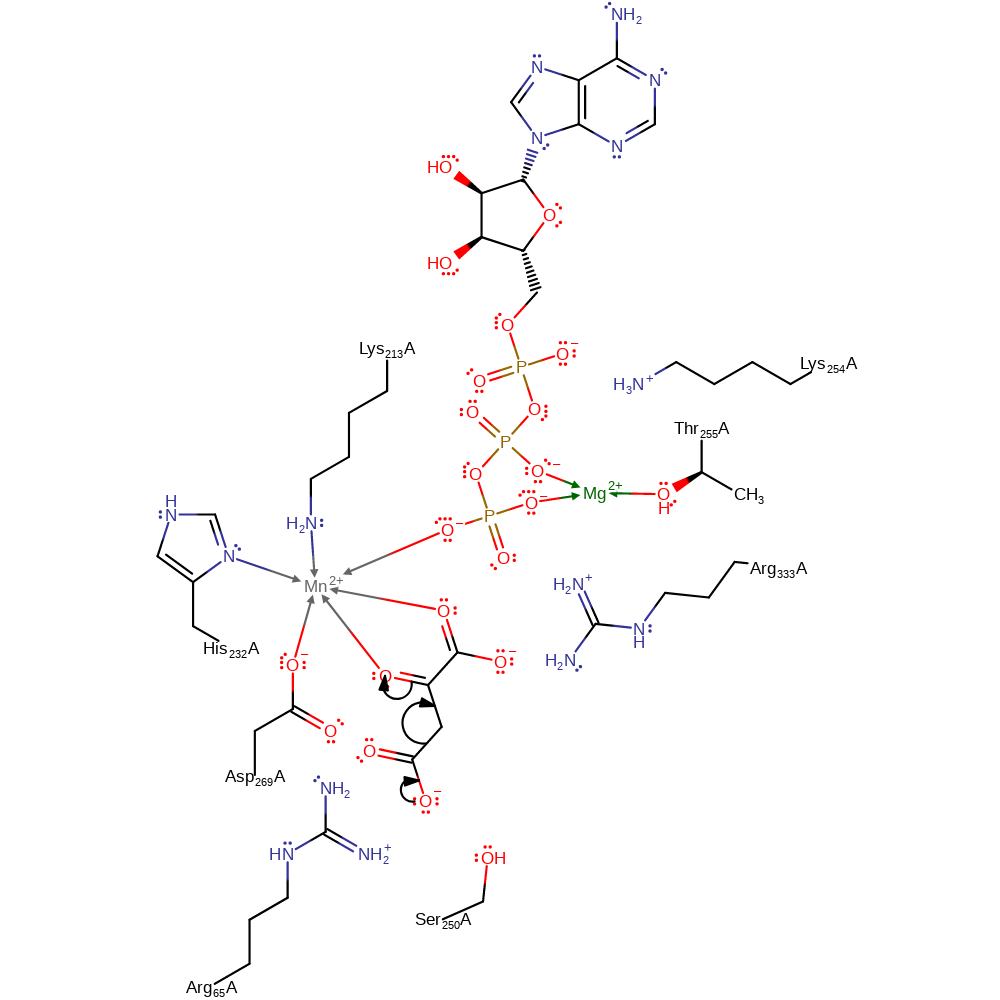
Step 1. The substrate undergoes decarboxylation, resulting in the formation of a enol-intermediate
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg333A | electrostatic stabiliser, hydrogen bond donor |
| His232A | hydrogen bond donor, electrostatic stabiliser |
| Lys254A | electrostatic stabiliser, hydrogen bond donor |
| Ser250A | steric role |
| His232A | metal ligand |
| Lys213A | metal ligand |
| Asp269A | metal ligand |
| Thr255A | metal ligand |
| Arg65A | electrostatic stabiliser |
Chemical Components
ingold: unimolecular elimination by the conjugate base, decarboxylation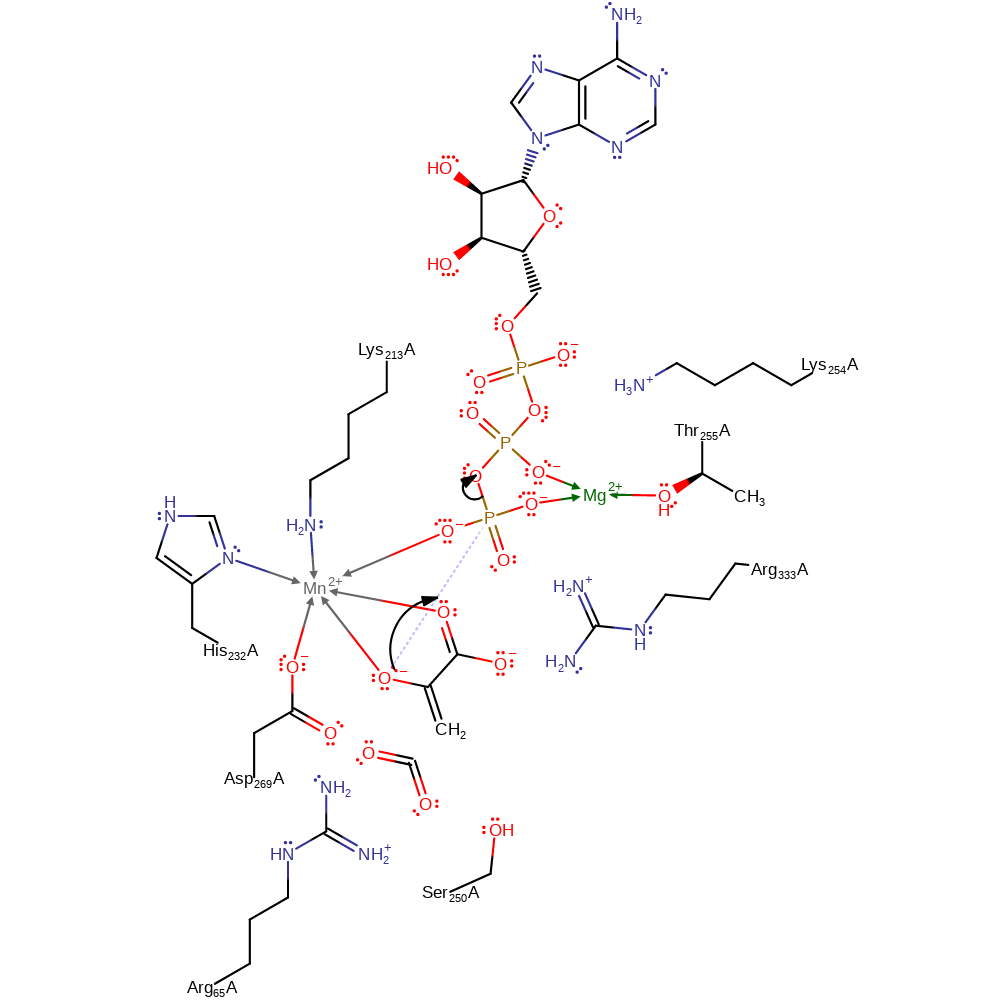
Step 2. The oxyanion formed attacks the gamma phosphate of ATP in a nucleophilic substitution reaction resulting in ADP and phosphoenolpyruvate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His232A | electrostatic stabiliser, hydrogen bond donor |
| Lys254A | electrostatic stabiliser, hydrogen bond donor |
| Arg333A | electrostatic stabiliser, hydrogen bond donor |
| Ser250A | steric role |
| His232A | metal ligand |
| Lys213A | metal ligand |
| Asp269A | metal ligand |
| Thr255A | metal ligand |
| Arg65A | increase electrophilicity |





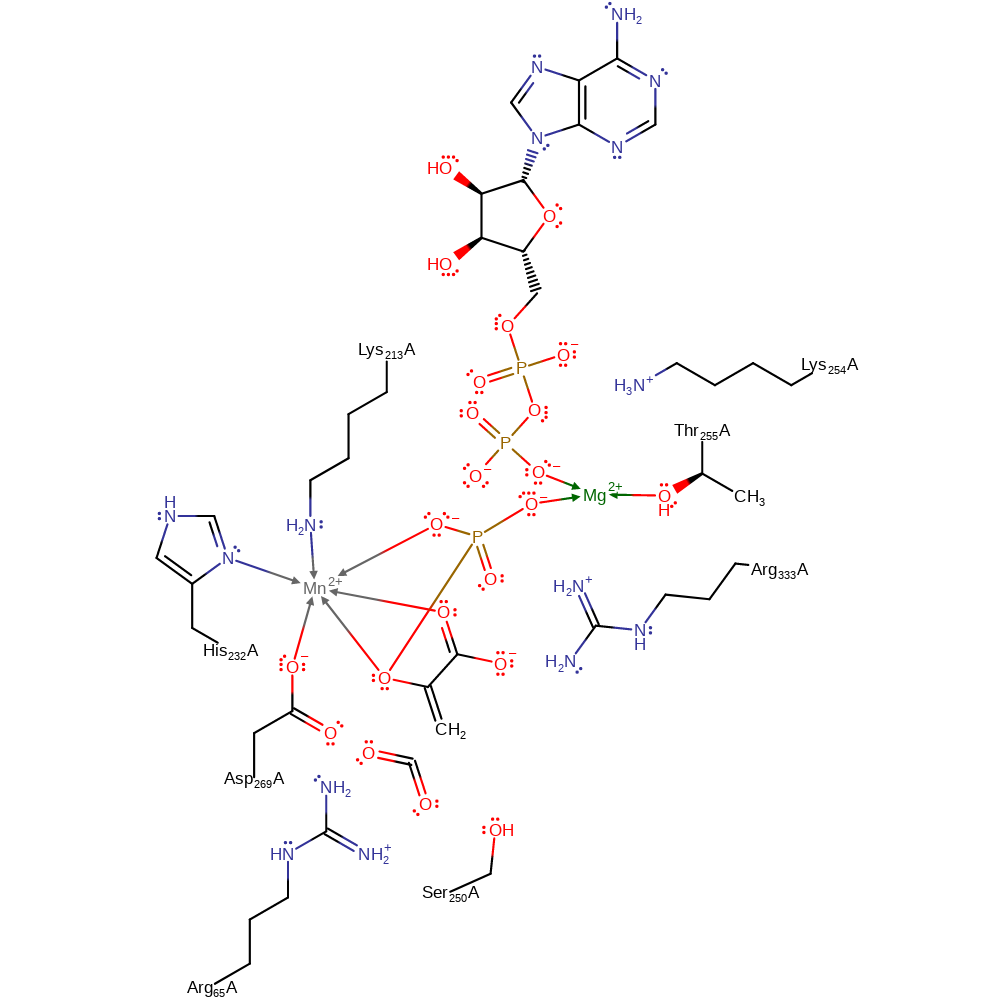 Download:
Download: