Glutathione-disulfide reductase
The pyrimidine nucleotide-disulphide oxidoreductases are a family of proteins which transfer electrons from NAD(P)H via FAD to a redox-active disulphide bond in the enzyme active site, which then reduces the substrate.
The structure of the ubiquitous enzyme glutathione reductase (EC 1.6.4.2), which helps protect cells from oxidative stress, is similar to trypanothione reductase (EC 1.6.4.8), lipoamide dehydrogenase (EC 1.8.1.4), higher eukaryotic thioredoxin reductase (EC 1.6.4.5) and mercuric reductase (1.16.1.1).
Reference Protein and Structure
- Sequence
-
P00390
 (1.8.1.7)
(1.8.1.7)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Homo sapiens (Human)

- PDB
-
2gh5
- Crystal Structure of human Glutathione Reductase complexed with a Fluoro-Analogue of the Menadione Derivative M5
(1.7 Å)



- Catalytic CATH Domains
-
3.30.390.30
 3.50.50.60
3.50.50.60  (see all for 2gh5)
(see all for 2gh5)
- Cofactors
- Fadh2(2-) (1), Glutathione (1)
Enzyme Reaction (EC:1.8.1.7)
Enzyme Mechanism
Introduction
Initially, the enzyme is in the oxidised state, with its redox active cysteines 58 and 63 forming a disulphide bond to each other. A hydride is transferred from NAD(P)H to FAD, a process facilitated by Glu201, Lys66 and Tyr197, as demonstrated by mutagenesis and structural studies. From there the electron makes an SN2 attack on the sulfur atom of Cys63, causing Cys58 to be displaced as thiolate. This residue is now ready to attack the substrate, which in all cases except mercuric reductase is another disulphide bond. His467 from the other subunit of the dimer has an essential role here, as shown by mutagenesis; first it seems to withdraw a proton from Cys58, activating the latter residue for a nucleophilic attack on the substrate. The proton is then poised to polarise the resulting Cys58-substrate mixed disulphide bond, making it vulnerable to attack from Cys63 to return the enzyme to its rest state. In addition to His467, another residue from the second subunit is found in the active site: Glu472 is shown to be essential by mutagenesis, and in structures is seen to orientate the catalytic Histidine.
Catalytic Residues Roles
| UniProt | PDB* (2gh5) | ||
| His511 | His467B | Acts as a general acid/base during the reaction. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Glu516 | Glu472B | Activates His467 | activator, hydrogen bond acceptor, electrostatic stabiliser |
| Cys102, Cys107 | Cys58A, Cys63A | These residues are in a disulfide bond at the start of the reaction and this bond provides the redox potential for the reduction reaction. | electrophile, electrofuge, nucleofuge, nucleophile |
| Tyr241, Lys110, Glu245 | Tyr197A, Lys66A, Glu201A | Helps facilitate the hydride transfer from NAD(P) to the FAD molecule. | activator |
Chemical Components
aromatic unimolecular elimination by the conjugate base, hydride transfer, aromatic bimolecular nucleophilic addition, cofactor used, intermediate formation, overall product formed, overall reactant used, rate-determining step, bimolecular nucleophilic substitution, enzyme-substrate complex formation, enzyme-substrate complex cleavage, intermediate collapse, intermediate terminated, native state of cofactor regenerated, proton transfer, native state of enzyme regeneratedReferences
- Deponte M (2013), Biochim Biophys Acta, 1830, 3217-3266. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. DOI:10.1016/j.bbagen.2012.09.018. PMID:23036594.
- Filomeni G et al. (2002), Biochem Pharmacol, 64, 1057-1064. Cell signalling and the glutathione redox system. DOI:10.1016/s0006-2952(02)01176-0.
- Sweet WL et al. (1991), Biochemistry, 30, 8702-8709. Human erythrocyte glutathione reductase: chemical mechanism and structure of the transition state for hydride transfer. DOI:10.1021/bi00099a031. PMID:1888731.
- Karplus PA et al. (1989), J Mol Biol, 210, 163-180. Substrate binding and catalysis by glutathione reductase as derived from refined enzyme: Substrate crystal structures at 2Å resolution. DOI:10.1016/0022-2836(89)90298-2. PMID:2585516.
- Deonarain MP et al. (1989), Biochemistry, 28, 9602-9607. Alternative proton donors/acceptors in the catalytic mechanism of the glutathione reductase of Escherichia coli: the role of histidine-439 and tyrosine-99. DOI:10.1021/bi00451a008. PMID:2558727.
- Karplus PA et al. (1987), J Mol Biol, 195, 701-729. Refined structure of glutathione reductase at 1.54 Å resolution. DOI:10.1016/0022-2836(87)90191-4. PMID:3656429.
- Pai EF et al. (1983), J Biol Chem, 258, 1752-1757. The catalytic mechanism of glutathione reductase as derived from x-ray diffraction analyses of reaction intermediates. PMID:6822532.
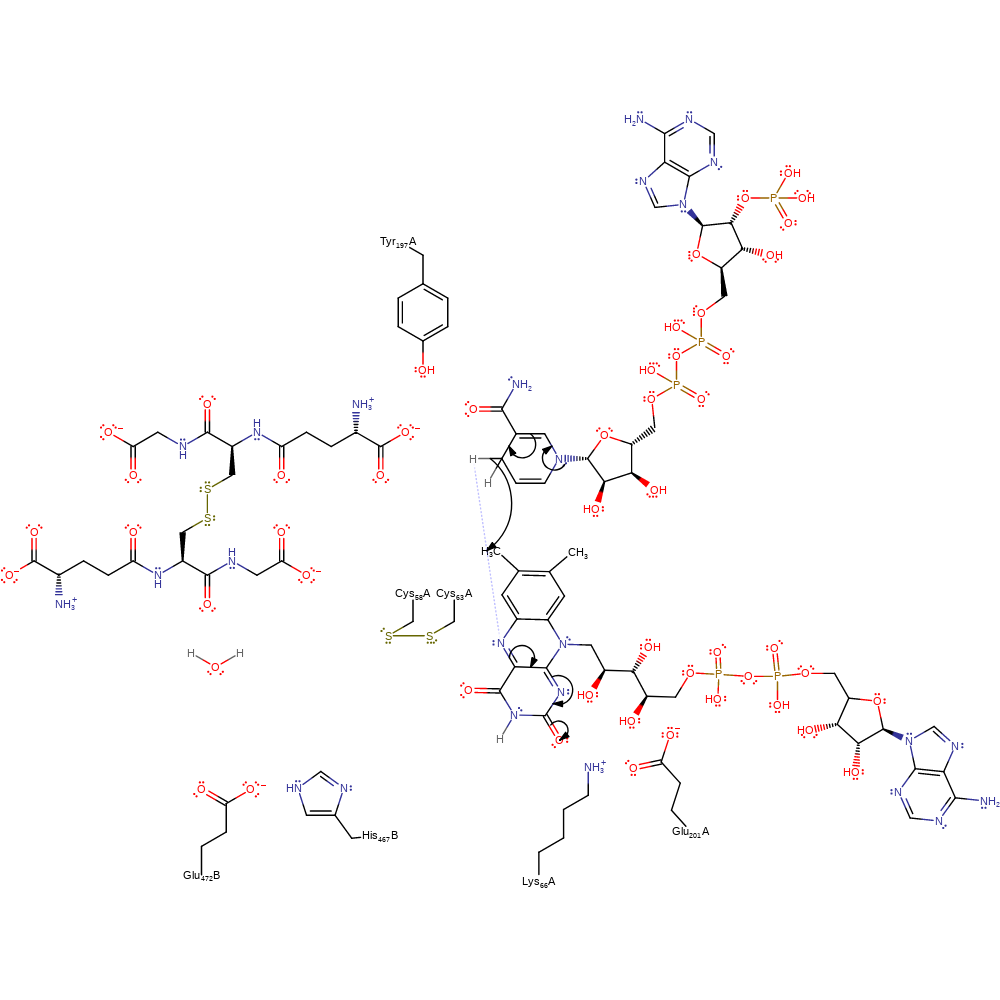
Step 1. NADP eliminates a hydride ion, which attacks the FAD cofactor with concomitant double bond rearrangement.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His467B | hydrogen bond donor |
| Lys66A | hydrogen bond donor |
| Glu472B | hydrogen bond acceptor |
| Glu201A | hydrogen bond acceptor, electrostatic stabiliser |
| Tyr197A | activator |
| Lys66A | activator |
| Glu201A | activator |
Chemical Components
ingold: aromatic unimolecular elimination by the conjugate base, hydride transfer, ingold: aromatic bimolecular nucleophilic addition, cofactor used, intermediate formation, overall product formed, overall reactant used, rate-determining step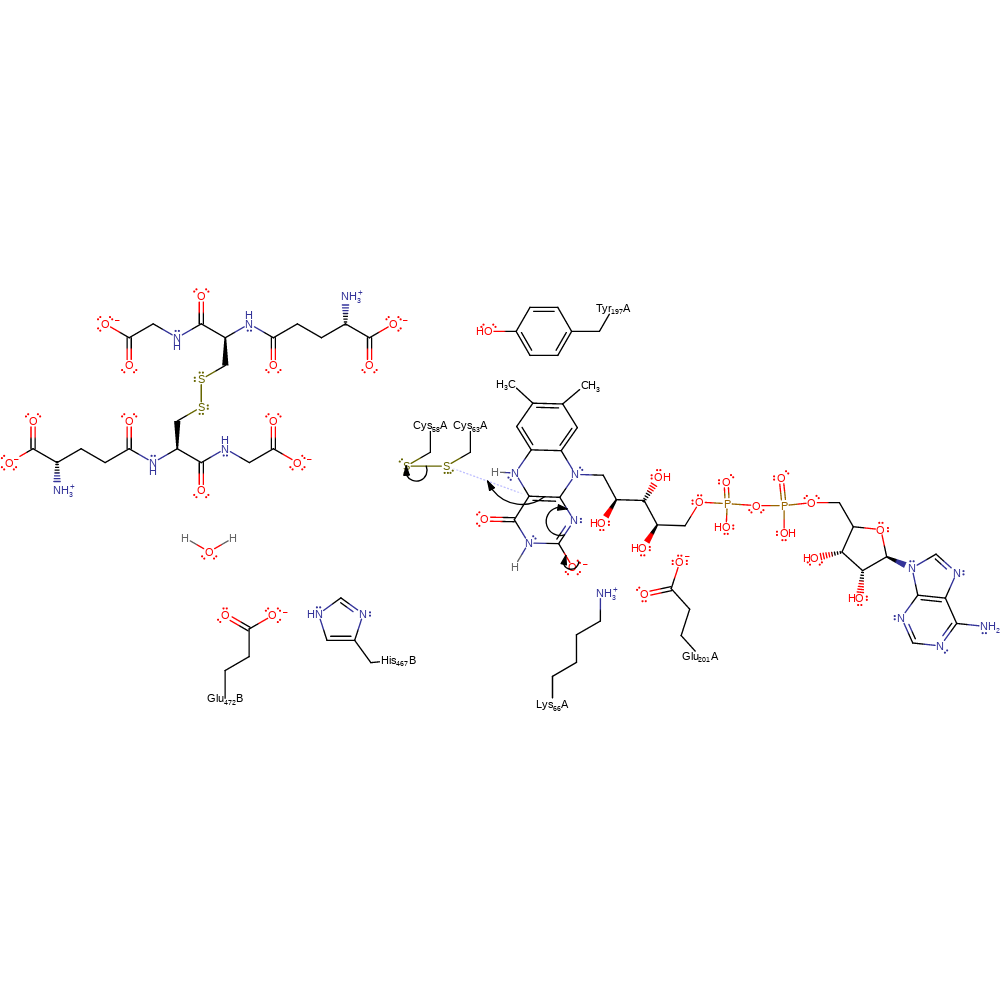
Step 2. The FAD oxyanion collapses, initiating a nucleophilic attack on Cys63 in a substitution reaction that eliminates Cys58 from the disulfide bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His467B | hydrogen bond donor |
| Lys66A | hydrogen bond donor, electrostatic stabiliser |
| Glu472B | hydrogen bond acceptor |
| Glu201A | hydrogen bond acceptor, electrostatic stabiliser |
| Cys58A | nucleofuge |
| Cys63A | electrofuge, electrophile |
Chemical Components
ingold: bimolecular nucleophilic substitution, enzyme-substrate complex formation, intermediate formation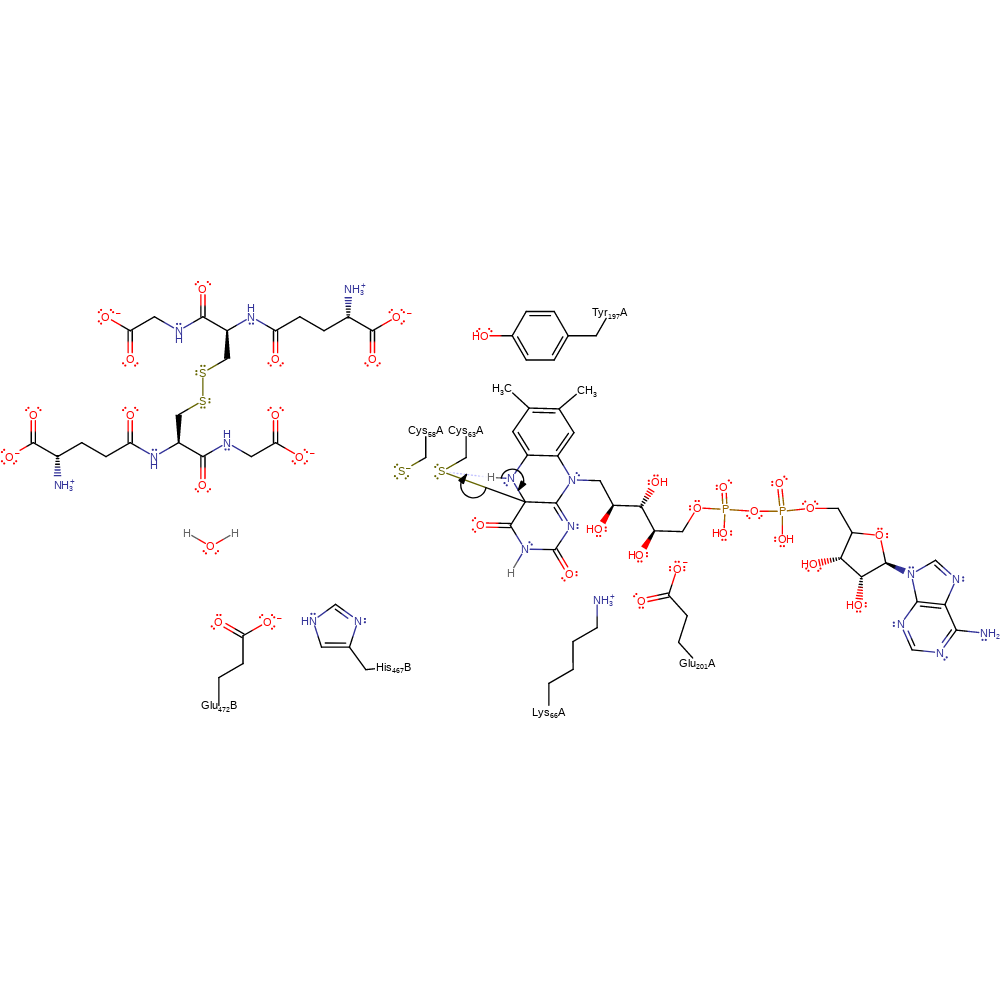
Step 3. The FAD cofactor eliminates Cys63 with concomitant deprotonation of the nitrogen to which the hydride was added.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys63A | hydrogen bond acceptor |
| His467B | hydrogen bond donor |
| Lys66A | hydrogen bond donor |
| Glu472B | hydrogen bond acceptor |
| Glu201A | hydrogen bond acceptor |
| Cys63A | nucleofuge, proton acceptor |
Chemical Components
ingold: aromatic unimolecular elimination by the conjugate base, enzyme-substrate complex cleavage, intermediate collapse, intermediate terminated, native state of cofactor regeneratedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys63A | hydrogen bond donor |
| His467B | hydrogen bond acceptor, hydrogen bond donor |
| Lys66A | hydrogen bond donor |
| Glu472B | hydrogen bond acceptor, activator |
| Glu201A | hydrogen bond acceptor |
| His467B | proton acceptor |
| Cys63A | proton donor |
Chemical Components
proton transfer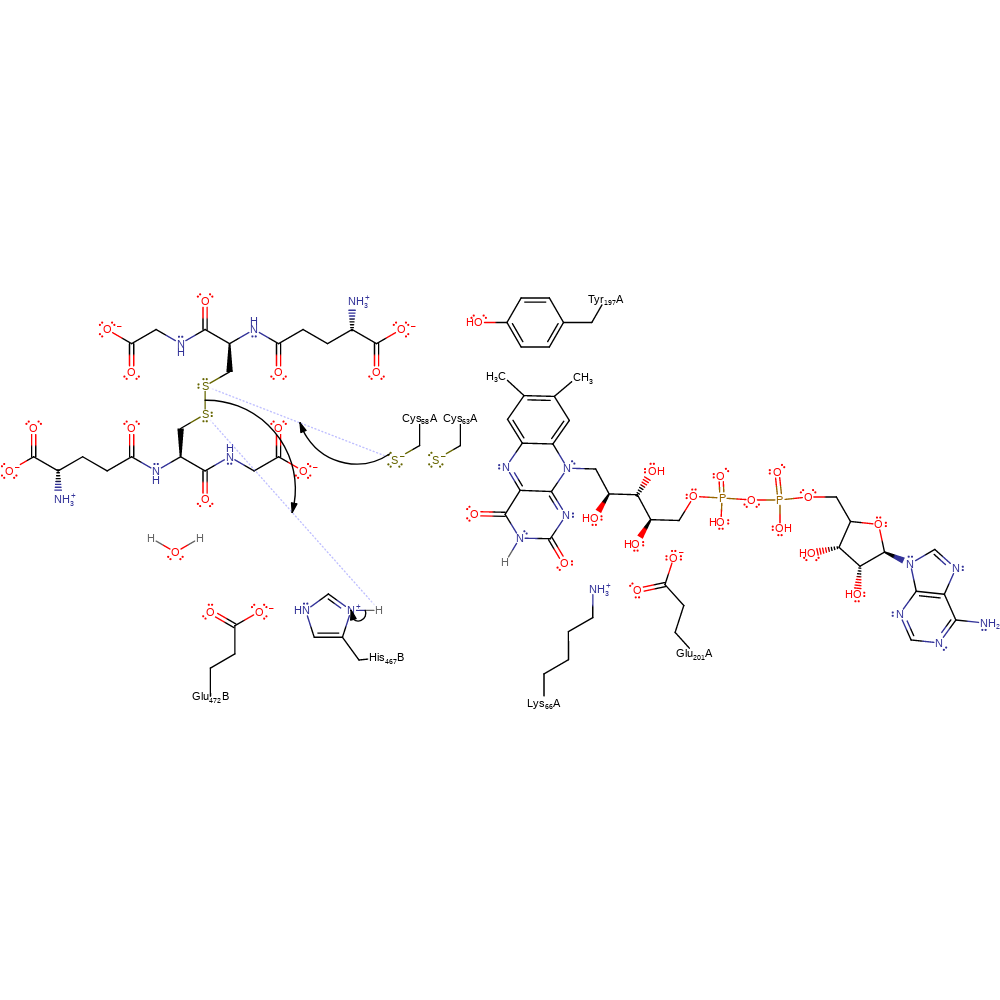
Step 5. Cys58 initiates a nucleophilic attack on the disulfide bond of oxidised glutathione in a substitution reaction, eliminating glutathione with concomitant deprotonation of His467B.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys66A | hydrogen bond donor |
| Glu201A | hydrogen bond acceptor |
| His467B | hydrogen bond donor |
| Glu472B | hydrogen bond acceptor, electrostatic stabiliser |
| Cys58A | nucleophile |
| His467B | proton donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, enzyme-substrate complex formation, intermediate formation, overall product formed, cofactor used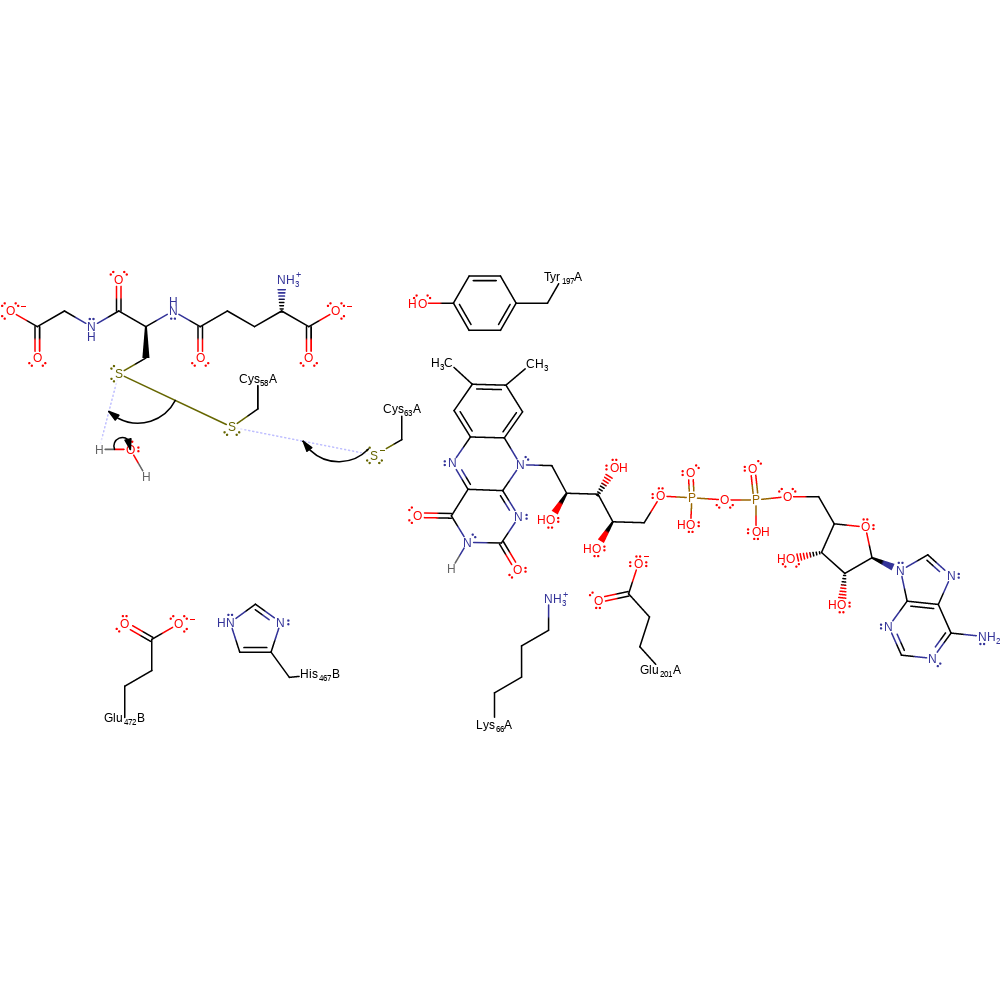
Step 6. Cys63 initiates a nucleophilic attack on Cys58 in a substitution reaction, eliminating glutathione with concomitant deprotonation of water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys66A | hydrogen bond donor |
| Glu201A | hydrogen bond acceptor |
| His467B | hydrogen bond donor, hydrogen bond acceptor |
| Glu472B | hydrogen bond acceptor |
| Cys58A | electrofuge, electrophile |
| Cys63A | nucleophile |






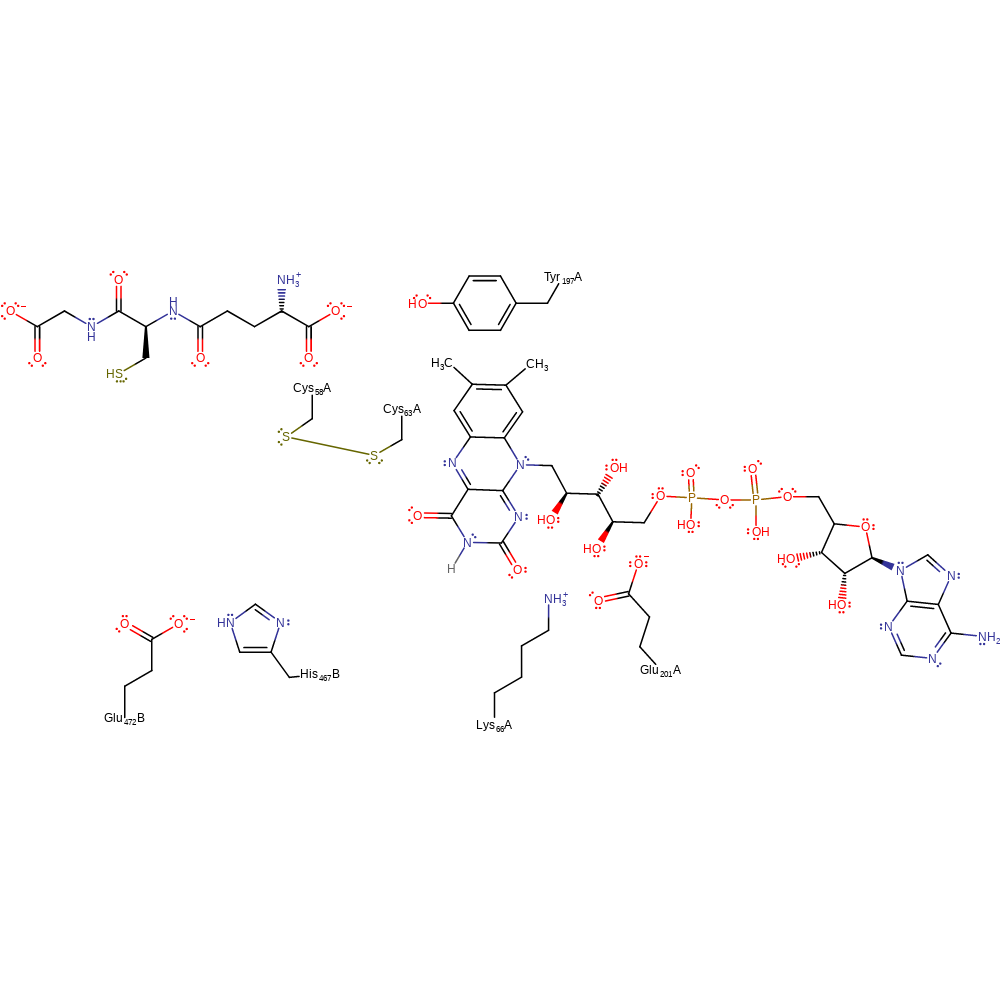 Download:
Download: