 |
PDBsum entry 9ofc

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Immune system, hydrolase
|
PDB id
|
|

|

|
9ofc
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Immune system, hydrolase
|
 |
|
Title:
|
 |
Cryoem structure of cad1 bound with ca4 and atp, hexamer with three intact dimers
|
|
Structure:
|
 |
Crispr-associated ring nuclease and adenosine deaminase, subunit a. Chain: a, b, g, h, k, l. Engineered: yes. Ca4 cleavage product, a2>p. Chain: c, d, i, j, m, n. Engineered: yes
|
|
Source:
|
 |
Thermoanaerobaculum aquaticum. Organism_taxid: 1312852. Gene: eg19_07865. Expressed in: escherichia coli. Expression_system_taxid: 562. Synthetic: yes. Organism_taxid: 1312852
|
|
Authors:
|
 |
Y.Zhao,H.Li
|
|
Key ref:
|
 |
M.Roth
et al.
Sequential conformational activation of a carf-Fused adenosine deaminase by cyclic oligoadenylates.
To be published,
.
PubMed id:
|
 |
|
Date:
|
 |
|
29-Apr-25
|
Release date:
|
20-Aug-25
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
A0A062XY19
(A0A062XY19_9BACT) -
adenosine deaminase from Thermoanaerobaculum aquaticum
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
625 a.a.
605 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.4.4
- adenosine deaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
adenosine + H2O + H+ = inosine + NH4+
|
|
2.
|
2'-deoxyadenosine + H2O + H+ = 2'-deoxyinosine + NH4+
|
|
 |
 |
 |
 |
 |
adenosine
Bound ligand (Het Group name = )
matches with 76.00% similarity
|
+
|
H2O
|
+
|
H(+)
|
=
|
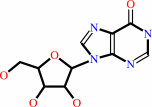 inosine
inosine
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
2'-deoxyadenosine
Bound ligand (Het Group name = )
matches with 72.00% similarity
|
+
|
H2O
|
+
|
H(+)
|
=
|
2'-deoxyinosine
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

