 |
PDBsum entry 2dub

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Enoyl-coa hydratase complexed with octanoyl-coa
|
|
Structure:
|
 |
2-enoyl-coa hydratase. Chain: a, b, c, d, e, f. Synonym: crotonase, enoyl-coa hydratase 1. Other_details: cocrystallized with octanoyl-coa
|
|
Source:
|
 |
Rattus norvegicus. Norway rat. Organism_taxid: 10116. Organ: liver. Cellular_location: mitochondria
|
|
Biol. unit:
|
 |
Hexamer (from PDB file)
|
|
Resolution:
|
 |
|
2.40Å
|
R-factor:
|
0.204
|
R-free:
|
0.260
|
|
|
Authors:
|
 |
C.K.Engel,R.K.Wierenga
|
Key ref:

|
 |
C.K.Engel
et al.
(1998).
The crystal structure of enoyl-CoA hydratase complexed with octanoyl-CoA reveals the structural adaptations required for binding of a long chain fatty acid-CoA molecule.
J Mol Biol,
275,
847-859.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
28-Apr-97
|
Release date:
|
29-Apr-98
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P14604
(ECHM_RAT) -
Enoyl-CoA hydratase, mitochondrial from Rattus norvegicus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
290 a.a.
254 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.4.2.1.17
- enoyl-CoA hydratase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a 4-saturated-(3S)-3-hydroxyacyl-CoA = a (3E)-enoyl-CoA + H2O
|
|
2.
|
a (3S)-3-hydroxyacyl-CoA = a (2E)-enoyl-CoA + H2O
|
|
 |
 |
 |
 |
 |
4-saturated-(3S)-3-hydroxyacyl-CoA
|
=
|
(3E)-enoyl-CoA
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
(3S)-3-hydroxyacyl-CoA
Bound ligand (Het Group name = )
matches with 88.14% similarity
|
=
|
(2E)-enoyl-CoA
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.5.3.3.8
- Delta(3)-Delta(2)-enoyl-CoA isomerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a (3Z)-enoyl-CoA = a 4-saturated (2E)-enoyl-CoA
|
|
2.
|
a (3E)-enoyl-CoA = a 4-saturated (2E)-enoyl-CoA
|
|
 |
 |
 |
 |
 |
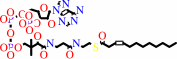 (3Z)-dodec-3-enoyl-CoA
(3Z)-dodec-3-enoyl-CoA
|
=
|
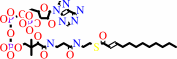 (2E)-dodec-2-enoyl-CoA
(2E)-dodec-2-enoyl-CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
275:847-859
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
The crystal structure of enoyl-CoA hydratase complexed with octanoyl-CoA reveals the structural adaptations required for binding of a long chain fatty acid-CoA molecule.
|
|
C.K.Engel,
T.R.Kiema,
J.K.Hiltunen,
R.K.Wierenga.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The structure of the hexameric rat mitochondrial enoyl-Coenzyme A (CoA)
hydratase, co-crystallised with the inhibitor octanoyl-CoA, has been refined at
a resolution of 2.4 A. Enoyl-CoA hydratase catalyses the hydration of
2,3-unsaturated enoyl-CoA thioesters. In the crystal structure only four of the
six active sites of the hexamer in the asymmetric unit are occupied with a
ligand molecule, showing an unliganded and a liganded active site within the
same crystal form. While the protein assembly and fold is identical to the
previously solved acetoacetyl-CoA complex, differences are observed close to the
fatty acid binding pocket due to the different nature of the ligands. The fatty
acid tail of octanoyl-CoA is bound in an extended conformation. This is possible
because a high B-factor loop, which separates in the acetoacetyl-CoA complex the
binding pocket of the acetoacetyl-CoA fatty acid tail from the intertrimer
space, has moved aside to allow binding of the longer octanoyl-CoA moiety. The
movement of this loop opens a tunnel which traverses the complete subunit from
the solvent space to the intertrimer space. The conformation of the catalytic
residues is identical, in both structures as well as in the liganded and the
unliganded active sites. In the unliganded active sites a water molecules is
bound between the two catalytic glutamate, residues Glu144 and Glu164. After
superposition of a liganded active site on an unliganded active site this water
molecule is close to the carbon centre that becomes hydroxylated in the
hydratase reaction. These findings support the idea that the active site is
rigid and that the catalytic residues and the water molecule, as seen in the
unliganded active site, are pre-positioned for very efficient catalysis.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. A, The reaction catalysed by enoyl-CoA hydratase. B, Covalent structure of the acetoacetyl moiety of the
inhibitor acetoacetyl-CoA. On the basis of spectroscopic measurements of acetoacetyl-CoA bound to hydratase it was
concluded that the 2,3-enolate form is the inhibitory species of the hydratase reaction (Waterson & Hill, 1972).
C, Covalent structure of the octanoyl moiety of the inhibitor octanoyl-CoA. D, Schematic drawing of Coenzyme A.
|
 |
Figure 5.
Figure 5. Stereo picture of the active site of the unliganded subunit D. Superimposed is the omit (Fo
-
Fc, aC density
from a model where the active site residues and the water molecule between them were removed. The distances
between this water molecule (X39) and the catalytic residues are indicated. The map is contoured at 3s.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(1998,
275,
847-859)
copyright 1998.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.M.Johnston,
V.L.Arcus,
and
E.N.Baker
(2005).
Structure of naphthoate synthase (MenB) from Mycobacterium tuberculosis in both native and product-bound forms.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
1199-1206.
|
 |
|
|
|
|
 |
M.C.Sleeman,
J.L.Sorensen,
E.T.Batchelar,
M.A.McDonough,
and
C.J.Schofield
(2005).
Structural and mechanistic studies on carboxymethylproline synthase (CarB), a unique member of the crotonase superfamily catalyzing the first step in carbapenem biosynthesis.
|
| |
J Biol Chem,
280,
34956-34965.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.A.Hubbard,
W.Yu,
H.Schulz,
and
J.J.Kim
(2005).
Domain swapping in the low-similarity isomerase/hydratase superfamily: the crystal structure of rat mitochondrial Delta3, Delta2-enoyl-CoA isomerase.
|
| |
Protein Sci,
14,
1545-1555.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Ishikawa,
D.Tsuchiya,
T.Oyama,
Y.Tsunaka,
and
K.Morikawa
(2004).
Structural basis for channelling mechanism of a fatty acid beta-oxidation multienzyme complex.
|
| |
EMBO J,
23,
2745-2754.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.K.Koski,
A.M.Haapalainen,
J.K.Hiltunen,
and
T.Glumoff
(2004).
A two-domain structure of one subunit explains unique features of eukaryotic hydratase 2.
|
| |
J Biol Chem,
279,
24666-24672.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.K.Hiltunen,
A.M.Mursula,
H.Rottensteiner,
R.K.Wierenga,
A.J.Kastaniotis,
and
A.Gurvitz
(2003).
The biochemistry of peroxisomal beta-oxidation in the yeast Saccharomyces cerevisiae.
|
| |
FEMS Microbiol Rev,
27,
35-64.
|
 |
|
|
|
|
 |
P.R.Hall,
Y.F.Wang,
R.E.Rivera-Hainaj,
X.Zheng,
M.Pustai-Carey,
P.R.Carey,
and
V.C.Yee
(2003).
Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core.
|
| |
EMBO J,
22,
2334-2347.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Hisano,
T.Tsuge,
T.Fukui,
T.Iwata,
K.Miki,
and
Y.Doi
(2003).
Crystal structure of the (R)-specific enoyl-CoA hydratase from Aeromonas caviae involved in polyhydroxyalkanoate biosynthesis.
|
| |
J Biol Chem,
278,
617-624.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.F.Bell,
J.Wu,
Y.Feng,
and
P.J.Tonge
(2001).
Involvement of glycine 141 in substrate activation by enoyl-CoA hydratase.
|
| |
Biochemistry,
40,
1725-1733.
|
 |
|
|
|
|
 |
J.F.Baker-Malcolm,
M.Lantz,
V.E.Anderson,
and
C.Thorpe
(2000).
Novel inactivation of enoyl-CoA hydratase via beta-elimination of 5, 6-dichloro-7,7,7-trifluoro-4-thia-5-heptenoyl-CoA.
|
| |
Biochemistry,
39,
12007-12018.
|
 |
|
|
|
|
 |
M.M.Benning,
T.Haller,
J.A.Gerlt,
and
H.M.Holden
(2000).
New reactions in the crotonase superfamily: structure of methylmalonyl CoA decarboxylase from Escherichia coli.
|
| |
Biochemistry,
39,
4630-4639.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Haller,
T.Buckel,
J.Rétey,
and
J.A.Gerlt
(2000).
Discovering new enzymes and metabolic pathways: conversion of succinate to propionate by Escherichia coli.
|
| |
Biochemistry,
39,
4622-4629.
|
 |
|
|
|
|
 |
H.Xiang,
L.Luo,
K.L.Taylor,
and
D.Dunaway-Mariano
(1999).
Interchange of catalytic activity within the 2-enoyl-coenzyme A hydratase/isomerase superfamily based on a common active site template.
|
| |
Biochemistry,
38,
7638-7652.
|
 |
|
|
|
|
 |
T.R.Kiema,
C.K.Engel,
W.Schmitz,
S.A.Filppula,
R.K.Wierenga,
and
J.K.Hiltunen
(1999).
Mutagenic and enzymological studies of the hydratase and isomerase activities of 2-enoyl-CoA hydratase-1.
|
| |
Biochemistry,
38,
2991-2999.
|
 |
|
|
|
|
 |
Y.M.Qin,
M.S.Marttila,
A.M.Haapalainen,
K.M.Siivari,
T.Glumoff,
and
J.K.Hiltunen
(1999).
Yeast peroxisomal multifunctional enzyme: (3R)-hydroxyacyl-CoA dehydrogenase domains A and B are required for optimal growth on oleic acid.
|
| |
J Biol Chem,
274,
28619-28625.
|
 |
|
|
|
|
 |
A.Gurvitz,
A.M.Mursula,
A.Firzinger,
B.Hamilton,
S.H.Kilpeläinen,
A.Hartig,
H.Ruis,
J.K.Hiltunen,
and
H.Rottensteiner
(1998).
Peroxisomal Delta3-cis-Delta2-trans-enoyl-CoA isomerase encoded by ECI1 is required for growth of the yeast Saccharomyces cerevisiae on unsaturated fatty acids.
|
| |
J Biol Chem,
273,
31366-31374.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

