 |
PDBsum entry 1s4d

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure analysis of the s-adenosyl-l-methionine dependent uroporphyrinogen-iii c-methyltransferase sumt
|
|
Structure:
|
 |
Uroporphyrin-iii c-methyltransferase. Chain: a, b, d, e, f, g, h, i, j, k, l, m. Synonym: s-adenosyl-l-methionine uroporphyrinogen-iii c- methyltransferase, urogen iii methylase, sumt, uroporphyrinogen iii methylase, urom. Engineered: yes
|
|
Source:
|
 |
Pseudomonas denitrificans. Organism_taxid: 43306. Gene: coba. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
 Dimer (from
)
Dimer (from
)
|
|
Resolution:
|
 |
|
2.70Å
|
R-factor:
|
0.215
|
R-free:
|
0.260
|
|
|
Authors:
|
 |
J.Vevodova,R.M.Graham,E.Raux,H.L.Schubert,D.I.Roper,A.A.Brindley, A.I.Scott,C.A.Roessner,N.P.J.Stamford,M.E.Stroupe,E.D.Getzoff, M.J.Warren,K.S.Wilson
|
Key ref:

|
 |
J.Vévodová
et al.
(2004).
Structure/function studies on a S-adenosyl-L-methionine-dependent uroporphyrinogen III C methyltransferase (SUMT), a key regulatory enzyme of tetrapyrrole biosynthesis.
J Mol Biol,
344,
419-433.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
16-Jan-04
|
Release date:
|
30-Nov-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P21631
(SUMT_SINSX) -
Uroporphyrinogen-III C-methyltransferase from Sinorhizobium sp
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
280 a.a.
256 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.1.1.107
- uroporphyrinogen-III C-methyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Corrin Biosynthesis (part 1)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
uroporphyrinogen III + 2 S-adenosyl-L-methionine = precorrin-2 + 2 S-adenosyl-L-homocysteine + H+
|
 |
 |
 |
 |
 |
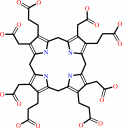 uroporphyrinogen III
uroporphyrinogen III
|
+
|
 2
×
S-adenosyl-L-methionine
2
×
S-adenosyl-L-methionine
|
=
|
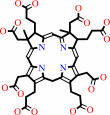 precorrin-2
precorrin-2
|
+
|
 2
×
S-adenosyl-L-homocysteine
2
×
S-adenosyl-L-homocysteine
|
+
|
H(+)
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
344:419-433
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure/function studies on a S-adenosyl-L-methionine-dependent uroporphyrinogen III C methyltransferase (SUMT), a key regulatory enzyme of tetrapyrrole biosynthesis.
|
|
J.Vévodová,
R.M.Graham,
E.Raux,
H.L.Schubert,
D.I.Roper,
A.A.Brindley,
A.Ian Scott,
C.A.Roessner,
N.P.Stamford,
M.Elizabeth Stroupe,
E.D.Getzoff,
M.J.Warren,
K.S.Wilson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystallographic structure of the Pseudomonas denitrificans
S-adenosyl-L-methionine-dependent uroporphyrinogen III methyltransferase (SUMT),
which is encoded by the cobA gene, has been solved by molecular replacement to
2.7A resolution. SUMT is a branchpoint enzyme that plays a key role in the
biosynthesis of modified tetrapyrroles by controlling flux to compounds such as
vitamin B(12) and sirohaem, and catalysing the transformation of
uroporphyrinogen III into precorrin-2. The overall topology of the enzyme is
similar to that of the SUMT module of sirohaem synthase (CysG) and the
cobalt-precorrin-4 methyltransferase CbiF and, as with the latter structures,
SUMT has the product S-adenosyl-L-homocysteine bound in the crystal. The roles
of a number of residues within the SUMT structure are discussed with respect to
their conservation either across the broader family of cobalamin biosynthetic
methyltransferases or within the sub-group of SUMT members. The D47N, L49A,
F106A, T130A, Y183A and M184A variants of SUMT were generated by mutagenesis of
the cobA gene, and tested for SAM binding and enzymatic activity. Of these
variants, only D47N and L49A bound the co-substrate S-adenosyl-L-methionine.
Consequently, all the mutants were severely restricted in their capacity to
synthesise precorrin-2, although both the D47N and L49A variants produced
significant quantities of precorrin-1, the monomethylated derivative of
uroporphyrinogen III. The activity of these variants is interpreted with respect
to the structure of the enzyme.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Figure 4. (a) Structure-based alignment of the sequences of
the transmethylase enzymes and domains from P. denitrificans
SUMT, Salmonella enterica CysG (multifunctional sirohaem
synthase) and Bacillus megaterium CbiF (anaerobic
cobalt-precorrin-4 methyltransferase) with the secondary
structure labelled. The 3D alignment was performed using the SSM
protein structure matching web page
(http://www.ebi.ac.uk/msd-srv/ssm). The residues indicated with
cyan dotted lines are either not modelled in the electron
density or excluded from the alignment by SSM as being too far
apart to be considered equivalent. (b) Superposition of the
SUMT, CysG and CbiF molecules including SAH. The position of the
ligand is almost identical in the three molecules. SUMT is
coloured cyan, CysG magenta and CbiF yellow (all including their
ligands).
|
 |
Figure 5.
Figure 5. (a) SAH binding by surrounding (2.6-3.8 Å)
residues is shown. Residues highlighted in the Figure interact
directly with SAH either by H-bonds or by van der Waals
contacts. (b) Molecule of SAH. 1s electron density from a 2F[o]
-F[c] map contoured in blue.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2004,
344,
419-433)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.Watanabe,
H.Oguri,
and
H.Oikawa
(2009).
Diversification of echinomycin molecular structure by way of chemoenzymatic synthesis and heterologous expression of the engineered echinomycin biosynthetic pathway.
|
| |
Curr Opin Chem Biol,
13,
189-196.
|
 |
|
|
|
|
 |
K.Watanabe,
K.Hotta,
A.P.Praseuth,
M.Searcey,
C.C.Wang,
H.Oguri,
and
H.Oikawa
(2009).
Rationally engineered total biosynthesis of a synthetic analogue of a natural quinomycin depsipeptide in Escherichia coli.
|
| |
Chembiochem,
10,
1965-1968.
|
 |
|
|
|
|
 |
R.S.Zajicek,
S.Bali,
S.Arnold,
A.A.Brindley,
M.J.Warren,
and
S.J.Ferguson
(2009).
d(1) haem biogenesis - assessing the roles of three nir gene products.
|
| |
FEBS J,
276,
6399-6411.
|
 |
|
|
|
|
 |
S.Storbeck,
J.Walther,
J.Müller,
V.Parmar,
H.M.Schiebel,
D.Kemken,
T.Dülcks,
M.J.Warren,
and
G.Layer
(2009).
The Pseudomonas aeruginosa nirE gene encodes the S-adenosyl-L-methionine-dependent uroporphyrinogen III methyltransferase required for heme d(1) biosynthesis.
|
| |
FEBS J,
276,
5973-5982.
|
 |
|
|
|
|
 |
G.L.Holliday,
J.M.Thornton,
A.Marquet,
A.G.Smith,
F.Rébeillé,
R.Mendel,
H.L.Schubert,
A.D.Lawrence,
and
M.J.Warren
(2007).
Evolution of enzymes and pathways for the biosynthesis of cofactors.
|
| |
Nat Prod Rep,
24,
972-987.
|
 |
|
|
|
|
 |
J.Fan,
Q.Liu,
Q.Hao,
M.Teng,
and
L.Niu
(2007).
Crystal structure of uroporphyrinogen decarboxylase from Bacillus subtilis.
|
| |
J Bacteriol,
189,
3573-3580.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Wada,
J.Harada,
Y.Yaeda,
H.Tamiaki,
H.Oh-Oka,
and
K.Fukuyama
(2007).
Crystal structures of CbiL, a methyltransferase involved in anaerobic vitamin B biosynthesis, and CbiL in complex with S-adenosylhomocysteine--implications for the reaction mechanism.
|
| |
FEBS J,
274,
563-573.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Frank,
E.Deery,
A.A.Brindley,
H.K.Leech,
A.Lawrence,
P.Heathcote,
H.L.Schubert,
K.Brocklehurst,
S.E.Rigby,
M.J.Warren,
and
R.W.Pickersgill
(2007).
Elucidation of substrate specificity in the cobalamin (vitamin B12) biosynthetic methyltransferases. Structure and function of the C20 methyltransferase (CbiL) from Methanothermobacter thermautotrophicus.
|
| |
J Biol Chem,
282,
23957-23969.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.Buchenau,
J.Kahnt,
I.U.Heinemann,
D.Jahn,
and
R.K.Thauer
(2006).
Heme biosynthesis in Methanosarcina barkeri via a pathway involving two methylation reactions.
|
| |
J Bacteriol,
188,
8666-8668.
|
 |
|
|
|
|
 |
P.H.Rehse,
T.Kitao,
and
T.H.Tahirov
(2005).
Structure of a closed-form uroporphyrinogen-III C-methyltransferase from Thermus thermophilus.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
913-919.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Z.Kozbial,
and
A.R.Mushegian
(2005).
Natural history of S-adenosylmethionine-binding proteins.
|
| |
BMC Struct Biol,
5,
19.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

