 |
PDBsum entry 1v9a

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.1.1.107
- uroporphyrinogen-III C-methyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Corrin Biosynthesis (part 1)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
uroporphyrinogen III + 2 S-adenosyl-L-methionine = precorrin-2 + 2 S-adenosyl-L-homocysteine + H+
|
 |
 |
 |
 |
 |
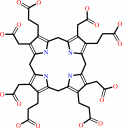 uroporphyrinogen III
uroporphyrinogen III
|
+
|
 2
×
S-adenosyl-L-methionine
2
×
S-adenosyl-L-methionine
|
=
|
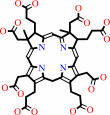 precorrin-2
precorrin-2
|
+
|
 2
×
S-adenosyl-L-homocysteine
2
×
S-adenosyl-L-homocysteine
|
+
|
H(+)
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
61:913-919
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of a closed-form uroporphyrinogen-III C-methyltransferase from Thermus thermophilus.
|
|
P.H.Rehse,
T.Kitao,
T.H.Tahirov.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Uroporphyrinogen-III C-methyltransferase from Thermus thermophilus is a
multifunctional protein responsible for two of the eight
S-adenosyl-methionine-dependent methylations of the corrin ring during vitamin
B(12) synthesis. The structure of this protein has been solved to 2.0 A
resolution in both the apo and cofactor-bound form. The monomer consists of two
domains, A and B, each consisting of a five-stranded beta-sheet and two or three
alpha-helices, with the cofactor bound at the interface. The biological unit is
the dimer found in the asymmetric unit. This dimer is related by a
non-crystallographic twofold such that two B domains combine to form a long
ten-stranded beta-sheet. When compared with solved related structures, this
structure shows clear differences in the region involved in cofactor and
substrate binding, affirming the role of several previously implicated residues
and questioning others. The solved related structures are characterized by an
exposed active site. The T. thermophilus structure has this site restricted by
the interaction of a flexible loop structure with a highly conserved residue,
suggesting a mechanistic role. This structure represents the ;closed' form of
the protein.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1
Structure of precorrin-2, the product of SAM-dependent uroporphyrinogen-III
C-methyltransferase. The two independent methylation sites are designated 1 and 2.
Methylation only at site 1 generates the intermediate precorrin-1.
|
 |
Figure 4.
Figure 4
Stereo diagram of the cofactor-binding site of uroporphyrinogen-III C-methyltransferase
from T. thermophilus (ttSUMT).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2005,
61,
913-919)
copyright 2005.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Y.Liao,
J.Deng,
A.Zhang,
M.Zhou,
Y.Hu,
H.Chen,
and
M.Jin
(2009).
Immunoproteomic analysis of outer membrane proteins and extracellular proteins of Actinobacillus pleuropneumoniae JL03 serotype 3.
|
| |
BMC Microbiol,
9,
172.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

