 |
PDBsum entry 1pd5

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of e.Coli chloramphenicol acetyltransferase type i at 2.5 angstrom resolution
|
|
Structure:
|
 |
Chloramphenicol acetyltransferase. Chain: a, b, c, d, e, f, g, h, i, j, k, l. Engineered: yes
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 562. Gene: cat or hcm1.206. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
 Trimer (from
)
Trimer (from
)
|
|
Resolution:
|
 |
|
2.50Å
|
R-factor:
|
0.199
|
R-free:
|
0.281
|
|
|
Authors:
|
 |
A.Roidis,M.Kokkinidis
|
|
Key ref:
|
 |
A.Roidis
and
m.kokkinidis
Crystal structure of e.Coli chloramphenicol acetyltransferase type i at 2.5 angstrom resolution.
To be published,
.
|
 |
|
Date:
|
 |
|
19-May-03
|
Release date:
|
01-Jun-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P62577
(CAT_ECOLX) -
Chloramphenicol acetyltransferase from Escherichia coli
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
219 a.a.
213 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.1.28
- chloramphenicol O-acetyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
chloramphenicol + acetyl-CoA = chloramphenicol 3-acetate + CoA
|
 |
 |
 |
 |
 |
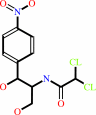 chloramphenicol
chloramphenicol
|
+
|
 acetyl-CoA
acetyl-CoA
|
=
|
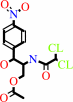 chloramphenicol 3-acetate
chloramphenicol 3-acetate
|
+
|
 CoA
CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

