 |
PDBsum entry 1oaf

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1oaf
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.11.1.11
- L-ascorbate peroxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-ascorbate + H2O2 = L-dehydroascorbate + 2 H2O
|
 |
 |
 |
 |
 |
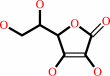 L-ascorbate
L-ascorbate
|
+
|
H2O2
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
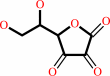 L-dehydroascorbate
L-dehydroascorbate
|
+
|
2
×
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Heme
|
 |
 |
 |
 |
 |
Heme
Bound ligand (Het Group name =
HEM)
matches with 95.45% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Struct Biol
10:303-307
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the ascorbate peroxidase-ascorbate complex.
|
|
K.H.Sharp,
M.Mewies,
P.C.Moody,
E.L.Raven.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Heme peroxidases catalyze the H2O2-dependent oxidation of a variety of
substrates, most of which are organic. Mechanistically, these enzymes are well
characterized: they share a common catalytic cycle that involves formation of a
two-electron, oxidized Compound I intermediate followed by two single-electron
reduction steps by substrate. The substrate specificity is more diverse--most
peroxidases oxidize small organic substrates, but there are prominent
exceptions--and there is a notable absence of structural information for a
representative peroxidase-substrate complex. Thus, the features that control
substrate specificity remain undefined. We present the structure of the complex
of ascorbate peroxidase-ascorbate. The structure defines the ascorbate-binding
interaction for the first time and provides new rationalization of the unusual
functional features of the related cytochrome c peroxidase enzyme, which has
been a benchmark for peroxidase catalysis for more than 20 years. A new
mechanism for electron transfer is proposed that challenges existing views of
substrate oxidation in other peroxidases.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. The active site of rsAPX32, 33. The key residues
are labeled, and water molecules are shown as red spheres.
Hydrogen bonds are indicated by green dotted lines.
|
 |
Figure 2.
Figure 2. The ascorbate-binding site. a, Stereo
representation of the overall structure of the rsAPX -ascorbate
complex34, showing the heme, the proximal and distal histidine
residues, the coordinated water molecule and the bound
ascorbate. The regions 20 -35 and 179 -181 (shown in Fig. 3) are
highlighted in magenta. b, The structure of L-ascorbic acid,
showing the L configuration at C^5. The pK[a]s of the 2-OH and
3-OH groups are 11.3 and 4.0, respectively35. c, The structure
of rsAPX showing the  -meso
and -meso
and  -meso
positions of the heme and bound solvent in the ascorbate-binding
site. d, Stereo view of the rsAPX -ascorbate complex, showing
refined electron density (green) and the binding of the
ascorbate. Hydrogen bonds are indicated (dotted lines)32, 33. -meso
positions of the heme and bound solvent in the ascorbate-binding
site. d, Stereo view of the rsAPX -ascorbate complex, showing
refined electron density (green) and the binding of the
ascorbate. Hydrogen bonds are indicated (dotted lines)32, 33.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nat Struct Biol
(2003,
10,
303-307)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.K.Singh,
R.P.Kumar,
N.Pandey,
N.Singh,
M.Sinha,
A.Bhushan,
P.Kaur,
S.Sharma,
and
T.P.Singh
(2010).
Mode of binding of the tuberculosis prodrug isoniazid to heme peroxidases: binding studies and crystal structure of bovine lactoperoxidase with isoniazid at 2.7 A resolution.
|
| |
J Biol Chem,
285,
1569-1576.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.K.Weng,
and
C.Chapple
(2010).
The origin and evolution of lignin biosynthesis.
|
| |
New Phytol,
187,
273-285.
|
 |
|
|
|
|
 |
T.Ishikawa,
N.Tajima,
H.Nishikawa,
Y.Gao,
M.Rapolu,
H.Shibata,
Y.Sawa,
and
S.Shigeoka
(2010).
Euglena gracilis ascorbate peroxidase forms an intramolecular dimeric structure: its unique molecular characterization.
|
| |
Biochem J,
426,
125-134.
|
 |
|
|
|
|
 |
A.K.Singh,
N.Singh,
M.Sinha,
A.Bhushan,
P.Kaur,
A.Srinivasan,
S.Sharma,
and
T.P.Singh
(2009).
Binding modes of aromatic ligands to mammalian heme peroxidases with associated functional implications: crystal structures of lactoperoxidase complexes with acetylsalicylic acid, salicylhydroxamic acid, and benzylhydroxamic acid.
|
| |
J Biol Chem,
284,
20311-20318.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.K.Singh,
N.Singh,
S.Sharma,
K.Shin,
M.Takase,
P.Kaur,
A.Srinivasan,
and
T.P.Singh
(2009).
Inhibition of lactoperoxidase by its own catalytic product: crystal structure of the hypothiocyanate-inhibited bovine lactoperoxidase at 2.3-A resolution.
|
| |
Biophys J,
96,
646-654.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
F.J.Ruiz-Dueñas,
M.Morales,
E.García,
Y.Miki,
M.J.Martínez,
and
A.T.Martínez
(2009).
Substrate oxidation sites in versatile peroxidase and other basidiomycete peroxidases.
|
| |
J Exp Bot,
60,
441-452.
|
 |
|
|
|
|
 |
C.Metcalfe,
I.K.Macdonald,
E.J.Murphy,
K.A.Brown,
E.L.Raven,
and
P.C.Moody
(2008).
The tuberculosis prodrug isoniazid bound to activating peroxidases.
|
| |
J Biol Chem,
283,
6193-6200.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Stjernschantz,
B.M.van Vugt-Lussenburg,
A.Bonifacio,
S.B.de Beer,
G.van der Zwan,
C.Gooijer,
J.N.Commandeur,
N.P.Vermeulen,
and
C.Oostenbrink
(2008).
Structural rationalization of novel drug metabolizing mutants of cytochrome P450 BM3.
|
| |
Proteins,
71,
336-352.
|
 |
|
|
|
|
 |
N.Najami,
T.Janda,
W.Barriah,
G.Kayam,
M.Tal,
M.Guy,
and
M.Volokita
(2008).
Ascorbate peroxidase gene family in tomato: its identification and characterization.
|
| |
Mol Genet Genomics,
279,
171-182.
|
 |
|
|
|
|
 |
S.Kitajima,
M.Kurioka,
T.Yoshimoto,
M.Shindo,
K.Kanaori,
K.Tajima,
and
K.Oda
(2008).
A cysteine residue near the propionate side chain of heme is the radical site in ascorbate peroxidase.
|
| |
FEBS J,
275,
470-480.
|
 |
|
|
|
|
 |
T.Ishikawa,
and
S.Shigeoka
(2008).
Recent advances in ascorbate biosynthesis and the physiological significance of ascorbate peroxidase in photosynthesizing organisms.
|
| |
Biosci Biotechnol Biochem,
72,
1143-1154.
|
 |
|
|
|
|
 |
V.Guallar,
and
F.Wallrapp
(2008).
Mapping protein electron transfer pathways with QM/MM methods.
|
| |
J R Soc Interface,
5,
S233-S239.
|
 |
|
|
|
|
 |
K.Fukuyama,
and
T.Okada
(2007).
Structures of cyanide, nitric oxide and hydroxylamine complexes of Arthromyces ramosusperoxidase at 100 K refined to 1.3 A resolution: coordination geometries of the ligands to the haem iron.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
472-477.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Huang,
and
P.R.Ortiz de Montellano
(2007).
Arthromyces ramosus peroxidase produces two chlorinating species.
|
| |
Biochem Biophys Res Commun,
355,
581-586.
|
 |
|
|
|
|
 |
T.L.Poulos
(2007).
The Janus nature of heme.
|
| |
Nat Prod Rep,
24,
504-510.
|
 |
|
|
|
|
 |
E.Gelhaye,
N.Navrot,
I.K.Macdonald,
N.Rouhier,
E.L.Raven,
and
J.P.Jacquot
(2006).
Ascorbate peroxidase-thioredoxin interaction.
|
| |
Photosynth Res,
89,
193-200.
|
 |
|
|
|
|
 |
J.N.Harvey,
C.M.Bathelt,
and
A.J.Mulholland
(2006).
QM/MM modeling of compound I active species in cytochrome P450, cytochrome C peroxidase, and ascorbate peroxidase.
|
| |
J Comput Chem,
27,
1352-1362.
|
 |
|
|
|
|
 |
P.J.Linley,
M.Landsberger,
T.Kohchi,
J.B.Cooper,
and
M.J.Terry
(2006).
The molecular basis of heme oxygenase deficiency in the pcd1 mutant of pea.
|
| |
FEBS J,
273,
2594-2606.
|
 |
|
|
|
|
 |
S.K.Badyal,
M.G.Joyce,
K.H.Sharp,
H.E.Seward,
M.Mewies,
J.Basran,
I.K.Macdonald,
P.C.Moody,
and
E.L.Raven
(2006).
Conformational mobility in the active site of a heme peroxidase.
|
| |
J Biol Chem,
281,
24512-24520.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.M.Bathelt,
A.J.Mulholland,
and
J.N.Harvey
(2005).
QM/MM studies of the electronic structure of the compound I intermediate in cytochrome c peroxidase and ascorbate peroxidase.
|
| |
Dalton Trans,
(),
3470-3476.
|
 |
|
|
|
|
 |
D.J.Stuehr,
C.C.Wei,
Z.Wang,
and
R.Hille
(2005).
Exploring the redox reactions between heme and tetrahydrobiopterin in the nitric oxide synthases.
|
| |
Dalton Trans,
(),
3427-3435.
|
 |
|
|
|
|
 |
F.K.Teixeira,
L.Menezes-Benavente,
R.Margis,
and
M.Margis-Pinheiro
(2004).
Analysis of the molecular evolutionary history of the ascorbate peroxidase gene family: inferences from the rice genome.
|
| |
J Mol Evol,
59,
761-770.
|
 |
|
|
|
|
 |
M.Zámocký
(2004).
Phylogenetic relationships in class I of the superfamily of bacterial, fungal, and plant peroxidases.
|
| |
Eur J Biochem,
271,
3297-3309.
|
 |
|
|
|
|
 |
R.Pierattelli,
L.Banci,
N.A.Eady,
J.Bodiguel,
J.N.Jones,
P.C.Moody,
E.L.Raven,
B.Jamart-Grégoire,
and
K.A.Brown
(2004).
Enzyme-catalyzed mechanism of isoniazid activation in class I and class III peroxidases.
|
| |
J Biol Chem,
279,
39000-39009.
|
 |
|
|
|
|
 |
T.Bertrand,
N.A.Eady,
J.N.Jones,
Jesmin,
J.M.Nagy,
B.Jamart-Grégoire,
E.L.Raven,
and
K.A.Brown
(2004).
Crystal structure of Mycobacterium tuberculosis catalase-peroxidase.
|
| |
J Biol Chem,
279,
38991-38999.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Z.Zhang,
J.S.Ren,
I.J.Clifton,
and
C.J.Schofield
(2004).
Crystal structure and mechanistic implications of 1-aminocyclopropane-1-carboxylic acid oxidase--the ethylene-forming enzyme.
|
| |
Chem Biol,
11,
1383-1394.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

