 |
PDBsum entry 2ghh

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2ghh
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.11.1.11
- L-ascorbate peroxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-ascorbate + H2O2 = L-dehydroascorbate + 2 H2O
|
 |
 |
 |
 |
 |
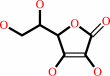 L-ascorbate
L-ascorbate
|
+
|
 H2O2
H2O2
|
=
|
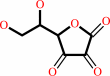 L-dehydroascorbate
L-dehydroascorbate
|
+
|
2
×
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Heme
|
 |
 |
 |
 |
 |
Heme
Bound ligand (Het Group name =
HEM)
matches with 95.45% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
281:24512-24520
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Conformational mobility in the active site of a heme peroxidase.
|
|
S.K.Badyal,
M.G.Joyce,
K.H.Sharp,
H.E.Seward,
M.Mewies,
J.Basran,
I.K.Macdonald,
P.C.Moody,
E.L.Raven.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Conformational mobility of the distal histidine residue has been implicated for
several different heme peroxidase enzymes, but unambiguous structural evidence
is not available. In this work, we present mechanistic, spectroscopic, and
structural evidence for peroxide- and ligand-induced conformational mobility of
the distal histidine residue (His-42) in a site-directed variant of ascorbate
peroxidase (W41A). In this variant, His-42 binds "on" to the heme in
the oxidized form, duplicating the active site structure of the cytochromes b
but, in contrast to the cytochromes b, is able to swing "off" the iron
during catalysis. This conformational flexibility between the on and off forms
is fully reversible and is used as a means to overcome the inherently unreactive
nature of the on form toward peroxide, so that essentially complete catalytic
activity is maintained. Contrary to the widely adopted view of heme enzyme
catalysis, these data indicate that strong coordination of the distal histidine
to the heme iron does not automatically undermine catalytic activity. The data
add a new dimension to our wider appreciation of structure/activity correlations
in other heme enzymes.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
FIGURE 4. A, the active site of ferric W41A, showing the
coordination of His-42 to the iron. SigmaA-weighted 2F[o] - F[c]
electron density at 1  is shown in blue, and
sigmaA-weighted F[o] - F[c] electron density at 3 is shown in blue, and
sigmaA-weighted F[o] - F[c] electron density at 3  is shown
in green. The positive F[o] - F[c] electron density in green
overlays with the position of His-42 in the structure of rsAPX.
Water molecules are shown as red spheres. B, stereo view of a
structural alignment of the orientation of His-42 in rsAPX (in
blue, Protein Data Bank code 1OAG) with the active site in W41A
(in green). Water molecules are shown as red spheres for W41A.
This figure was created using PyMOL (40). is shown
in green. The positive F[o] - F[c] electron density in green
overlays with the position of His-42 in the structure of rsAPX.
Water molecules are shown as red spheres. B, stereo view of a
structural alignment of the orientation of His-42 in rsAPX (in
blue, Protein Data Bank code 1OAG) with the active site in W41A
(in green). Water molecules are shown as red spheres for W41A.
This figure was created using PyMOL (40).
|
 |
Figure 7.
FIGURE 7. Overlay of the structures of ferric W41A (green)
and ferric W41A after reaction with H[2]O[2] (yellow). Water
molecules in the two structures are shown in green and yellow,
respectively. The orientation of His-42 after reaction with
H[2]O[2] (yellow) overlays with that of rsAPX (see Fig. 4B).
This figure was created using PyMOL (40).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2006,
281,
24512-24520)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.Fukuyama,
and
T.Okada
(2007).
Structures of cyanide, nitric oxide and hydroxylamine complexes of Arthromyces ramosusperoxidase at 100 K refined to 1.3 A resolution: coordination geometries of the ligands to the haem iron.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
472-477.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Kitajima,
T.Shimaoka,
M.Kurioka,
and
A.Yokota
(2007).
Irreversible cross-linking of heme to the distal tryptophan of stromal ascorbate peroxidase in response to rapid inactivation by H2O2.
|
| |
FEBS J,
274,
3013-3020.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

